HLA class I molecules reflect an altered host proteome after influenza virus infection
- PMID: 19748539
- PMCID: PMC2795087
- DOI: 10.1016/j.humimm.2009.08.012
HLA class I molecules reflect an altered host proteome after influenza virus infection
Abstract
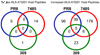
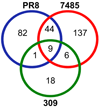
Class I HLA sample and display peptides from thousands of endogenous proteins at the cell surface. During infection, the influenza virus modifies the host cell proteome by triggering host antiviral responses, hijacking host processes, and inhibiting host mRNA processing. In turn, the catalog of HLA class I peptides that decorate the surface of an infected cell is positioned to reflect an altered host cell proteome. To understand the host-encoded peptides presented by class I molecules after influenza infection, we compared by mass spectrometry (MS) the peptides eluted from the HLA of naive and infected cells. We identified 20 peptide ligands unique to infected cells and 347 peptides with increased presentation after infection. Infection with different influenza strains demonstrated that proteome changes are predominantly strain-specific, with few individual cellular interactions observed for multiple viral strains. Modeling by pathway analysis, however, revealed that strain specific host peptide changes represent different routes to the same destination; host changes mediated by influenza are found predominantly clustered around HLA-B, ACTB, HSP90AB1, CDK2, and ANXA2. The class I HLA proteome scanning of influenza-infected cells therefore indicates how divergent strains of influenza pursue alternate routes to access the same host cell processes.
Figures

Similar articles
-
HLA class I molecules consistently present internal influenza epitopes.Proc Natl Acad Sci U S A. 2009 Jan 13;106(2):540-5. doi: 10.1073/pnas.0811271106. Epub 2009 Jan 2. Proc Natl Acad Sci U S A. 2009. PMID: 19122146 Free PMC article.
-
Downregulation of MHC Class I Expression by Influenza A and B Viruses.Front Immunol. 2019 May 29;10:1158. doi: 10.3389/fimmu.2019.01158. eCollection 2019. Front Immunol. 2019. PMID: 31191533 Free PMC article.
-
Acid Stripping after Infection Improves the Detection of Viral HLA Class I Natural Ligands Identified by Mass Spectrometry.Int J Mol Sci. 2021 Sep 29;22(19):10503. doi: 10.3390/ijms221910503. Int J Mol Sci. 2021. PMID: 34638844 Free PMC article.
-
HIV-1 proteins in infected cells determine the presentation of viral peptides by HLA class I and class II molecules and the nature of the cellular and humoral antiviral immune responses--a review.Virus Genes. 1994 Jul;8(3):249-70. doi: 10.1007/BF01704519. Virus Genes. 1994. PMID: 7975271 Review.
-
Direct class I HLA antigen discovery to distinguish virus-infected and cancerous cells.Expert Rev Proteomics. 2006 Dec;3(6):641-52. doi: 10.1586/14789450.3.6.641. Expert Rev Proteomics. 2006. PMID: 17181478 Review.
Cited by
-
Know thy immune self and non-self: Proteomics informs on the expanse of self and non-self, and how and where they arise.Proteomics. 2021 Dec;21(23-24):e2000143. doi: 10.1002/pmic.202000143. Epub 2021 Aug 9. Proteomics. 2021. PMID: 34310018 Free PMC article. Review.
-
Comprehensive Analysis of the Naturally Processed Peptide Repertoire: Differences between HLA-A and B in the Immunopeptidome.PLoS One. 2015 Sep 16;10(9):e0136417. doi: 10.1371/journal.pone.0136417. eCollection 2015. PLoS One. 2015. PMID: 26375851 Free PMC article.
-
Exploiting non-canonical translation to identify new targets for T cell-based cancer immunotherapy.Cell Mol Life Sci. 2018 Feb;75(4):607-621. doi: 10.1007/s00018-017-2628-4. Epub 2017 Aug 19. Cell Mol Life Sci. 2018. PMID: 28823056 Free PMC article. Review.
-
Position 45 influences the peptide binding motif of HLA-B*44:08.Immunogenetics. 2012 Mar;64(3):245-9. doi: 10.1007/s00251-011-0583-z. Epub 2011 Oct 19. Immunogenetics. 2012. PMID: 22009320
-
Defining the HLA class I-associated viral antigen repertoire from HIV-1-infected human cells.Eur J Immunol. 2016 Jan;46(1):60-9. doi: 10.1002/eji.201545890. Epub 2015 Nov 4. Eur J Immunol. 2016. PMID: 26467324 Free PMC article.
References
-
- Hickman HD, Luis AD, Buchli R, Few SR, Sathiamurthy M, VanGundy RS, et al. Toward a definition of self: proteomic evaluation of the class I Peptide repertoire. J Immunol. 2004 Mar 1;172(5):2944–2952. - PubMed
-
- Yewdell J, Bennink JR, Hosaka Y. Cells process exogenous proteins for recognition by cytotoxic T lymphocytes. Science. 1988;239:637–640. - PubMed
-
- Kash JC, Goodman AG, Korth MJ, Katze MG. Hijacking of the host-cell response and translational control during influenza virus infection. Virus Res. 2006 Jul;119(1):111–120. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

