Fine-tuning of the unfolded protein response: Assembling the IRE1alpha interactome
- PMID: 19748352
- PMCID: PMC3101568
- DOI: 10.1016/j.molcel.2009.08.021
Fine-tuning of the unfolded protein response: Assembling the IRE1alpha interactome
Abstract
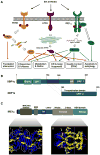
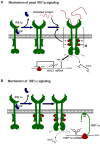
Endoplasmic reticulum (ER) stress is a hallmark feature of secretory cells and many diseases, including cancer, neurodegeneration, and diabetes. Adaptation to protein-folding stress is mediated by the activation of an integrated signal transduction pathway known as the unfolded protein response (UPR). The UPR signals through three distinct stress sensors located at the ER membrane-IRE1alpha, ATF6, and PERK. Although PERK and IRE1alpha share functionally similar ER-luminal sensing domains and both are simultaneously activated in cellular paradigms of ER stress in vitro, they are selectively engaged in vivo by the physiological stress of unfolded proteins. The differences in terms of tissue-specific regulation of the UPR may be explained by the formation of distinct regulatory protein complexes. This concept is supported by the recent identification of adaptor and modulator proteins that directly interact with IRE1alpha. In this Review, we discuss recent evidence supporting a model where IRE1alpha signaling emerges as a highly regulated process, controlled by the formation of a dynamic scaffold onto which many regulatory components assemble.
Figures
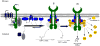
Similar articles
-
Endoplasmic reticulum stress in liver disease.J Hepatol. 2011 Apr;54(4):795-809. doi: 10.1016/j.jhep.2010.11.005. Epub 2010 Nov 13. J Hepatol. 2011. PMID: 21145844 Free PMC article. Review.
-
Phenformin activates the unfolded protein response in an AMP-activated protein kinase (AMPK)-dependent manner.J Biol Chem. 2013 May 10;288(19):13631-8. doi: 10.1074/jbc.M113.462762. Epub 2013 Apr 2. J Biol Chem. 2013. PMID: 23548904 Free PMC article.
-
Activation of alveolar epithelial ER stress by β-coronavirus infection disrupts surfactant homeostasis in mice: implications for COVID-19 respiratory failure.Am J Physiol Lung Cell Mol Physiol. 2024 Aug 1;327(2):L232-L249. doi: 10.1152/ajplung.00324.2023. Epub 2024 Jun 11. Am J Physiol Lung Cell Mol Physiol. 2024. PMID: 38860845
-
The Yin and Yang of hsa-miR-1244 expression levels during activation of the UPR control cell fate.Cell Commun Signal. 2024 Dec 2;22(1):577. doi: 10.1186/s12964-024-01967-2. Cell Commun Signal. 2024. PMID: 39623432 Free PMC article.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
Cited by
-
Sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) activity is required for V(D)J recombination.J Exp Med. 2021 Aug 2;218(8):e20201708. doi: 10.1084/jem.20201708. Epub 2021 May 25. J Exp Med. 2021. PMID: 34033676 Free PMC article.
-
Non-canonical function of IRE1α determines mitochondria-associated endoplasmic reticulum composition to control calcium transfer and bioenergetics.Nat Cell Biol. 2019 Jun;21(6):755-767. doi: 10.1038/s41556-019-0329-y. Epub 2019 May 20. Nat Cell Biol. 2019. PMID: 31110288 Free PMC article.
-
Brucella induces an unfolded protein response via TcpB that supports intracellular replication in macrophages.PLoS Pathog. 2013;9(12):e1003785. doi: 10.1371/journal.ppat.1003785. Epub 2013 Dec 5. PLoS Pathog. 2013. PMID: 24339776 Free PMC article.
-
Macrophage-Derived Immunoglobulin M Inhibits Inflammatory Responses via Modulating Endoplasmic Reticulum Stress.Cells. 2021 Oct 20;10(11):2812. doi: 10.3390/cells10112812. Cells. 2021. PMID: 34831038 Free PMC article.
-
The unfolded-protein-response sensor IRE-1α regulates the function of CD8α+ dendritic cells.Nat Immunol. 2014 Mar;15(3):248-57. doi: 10.1038/ni.2808. Epub 2014 Jan 19. Nat Immunol. 2014. PMID: 24441789
References
-
- Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, Lennon CJ, Kluger Y, Dynlacht BD. XBP1 Controls Diverse Cell Type- and Condition-Specific Transcriptional Regulatory Networks. Mol Cell. 2007;27:53–66. - PubMed
-
- Bailly-Maitre B, Fondevila C, Kaldas F, Droin N, Luciano F, Ricci JE, Croxton R, Krajewska M, Zapata JM, Kupiec-Weglinski JW, Farmer D, Reed JC. Cytoprotective gene bi-1 is required for intrinsic protection from endoplasmic reticulum stress and ischemia-reperfusion injury. Proc Natl Acad Sci U S A. 2006;103:2809–2814. - PMC - PubMed
-
- Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

