Mechanism of the internal ribosome entry site-mediated translation of serine hydroxymethyltransferase 1
- PMID: 19734143
- PMCID: PMC2781508
- DOI: 10.1074/jbc.M109.035576
Mechanism of the internal ribosome entry site-mediated translation of serine hydroxymethyltransferase 1
Abstract
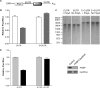
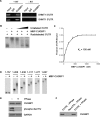
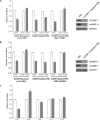
The 5'-untranslated region (UTR) of serine hydroxymethyltransferase 1 (SHMT1) contains an internal ribosome entry site (IRES) that regulates SHMT1 expression, a rate-limiting enzyme in de novo thymidylate biosynthesis. In this study, we show that the SHMT1 IRES is the first example of a cellular IRES that is poly(A) tail-independent. Interactions between the 5'-UTR and 3'-UTR functionally replaced interactions between the poly(A) tail and the poly(A)-binding protein (PABP) to achieve maximal IRES-mediated translational efficiency. Depletion of the SHMT1 IRES-specific trans-acting factor (ITAF) CUG-binding protein 1 (CUGBP1) from in vitro translation extracts or deletion of the CUGBP1 binding site on the 3'-UTR of the SHMT1 transcript decreased the IRES activity of non-polyadenylylated biscistronic mRNAs relative to polyadenylylated biscistronic mRNAs and resulted in a requirement for PABP. We also identified a novel ITAF, heterogeneous nuclear ribonucleoprotein H2 (hnRNP H2), that stimulates SHMT1 IRES activity by binding to the 5'-UTR of the transcript and interacting with CUGBP1. Collectively, these data support a model for the IRES-mediated translation of SHMT1 whereby the circularization of the mRNA typically provided by the eukaryotic initiation factor (eIF) 4G/PABP/poly(A) tail interaction is achieved instead through the hnRNP H2/CUGBP1-mediated interaction of the 5'- and 3'-UTRs of the SHMT1 transcript. This circularization enhances the IRES activity of SHMT1 by facilitating the recruitment and/or recycling of ribosomal subunits, which bind to the transcript in the middle of the 5'-UTR and migrate to the initiation codon via eIF4A-mediated scanning.
Figures
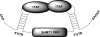


Similar articles
-
A UV-responsive internal ribosome entry site enhances serine hydroxymethyltransferase 1 expression for DNA damage repair.J Biol Chem. 2009 Nov 6;284(45):31097-108. doi: 10.1074/jbc.M109.015800. Epub 2009 Sep 4. J Biol Chem. 2009. PMID: 19734144 Free PMC article.
-
UNR translation can be driven by an IRES element that is negatively regulated by polypyrimidine tract binding protein.Nucleic Acids Res. 2005 May 31;33(10):3095-108. doi: 10.1093/nar/gki611. Print 2005. Nucleic Acids Res. 2005. PMID: 15928332 Free PMC article.
-
A ferritin-responsive internal ribosome entry site regulates folate metabolism.J Biol Chem. 2007 Oct 12;282(41):29927-35. doi: 10.1074/jbc.M706264200. Epub 2007 Aug 16. J Biol Chem. 2007. PMID: 17702748
-
Hepatitis C Virus Translation Regulation.Int J Mol Sci. 2020 Mar 27;21(7):2328. doi: 10.3390/ijms21072328. Int J Mol Sci. 2020. PMID: 32230899 Free PMC article. Review.
-
An atypical IRES within the 5' UTR of a dicistrovirus genome.Virus Res. 2009 Feb;139(2):157-65. doi: 10.1016/j.virusres.2008.07.017. Epub 2008 Sep 11. Virus Res. 2009. PMID: 18755228 Review.
Cited by
-
Linking Α to Ω: diverse and dynamic RNA-based mechanisms to regulate gene expression by 5'-to-3' communication.F1000Res. 2016 Aug 19;5:F1000 Faculty Rev-2037. doi: 10.12688/f1000research.7913.1. eCollection 2016. F1000Res. 2016. PMID: 27610229 Free PMC article. Review.
-
Targeting nuclear thymidylate biosynthesis.Mol Aspects Med. 2017 Feb;53:48-56. doi: 10.1016/j.mam.2016.11.005. Epub 2016 Nov 19. Mol Aspects Med. 2017. PMID: 27876557 Free PMC article. Review.
-
CELFish ways to modulate mRNA decay.Biochim Biophys Acta. 2013 Jun-Jul;1829(6-7):695-707. doi: 10.1016/j.bbagrm.2013.01.001. Epub 2013 Jan 15. Biochim Biophys Acta. 2013. PMID: 23328451 Free PMC article. Review.
-
Repeat-Associated Non-ATG Translation in Neurological Diseases.Cold Spring Harb Perspect Biol. 2018 Dec 3;10(12):a033019. doi: 10.1101/cshperspect.a033019. Cold Spring Harb Perspect Biol. 2018. PMID: 29891563 Free PMC article. Review.
-
Translational Regulation by hnRNP H/F Is Essential for the Proliferation and Survival of Glioblastoma.Cancers (Basel). 2022 Mar 2;14(5):1283. doi: 10.3390/cancers14051283. Cancers (Basel). 2022. PMID: 35267591 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

