Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid
- PMID: 19729508
- PMCID: PMC2770672
- DOI: 10.1093/nar/gkp707
Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid
Abstract
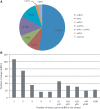
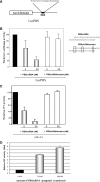
Small non-coding RNAs of 18-25 nt in length can regulate gene expression through the RNA interference (RNAi) pathway. To characterize small RNAs in HIV-1-infected cells, we performed linker-ligated cloning followed by high-throughput pyrosequencing. Here, we report the composition of small RNAs in HIV-1 productively infected MT4 T-cells. We identified several HIV-1 small RNA clones and a highly abundant small 18-nt RNA that is antisense to the HIV-1 primer-binding site (PBS). This 18-nt RNA apparently originated from the dsRNA hybrid formed by the HIV-1 PBS and the 3' end of the human cellular tRNAlys3. It was found to associate with the Ago2 protein, suggesting its possible function in the cellular RNAi machinery for targeting HIV-1.
Figures

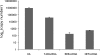


Similar articles
-
Minimal sequence requirements of a functional human immunodeficiency virus type 1 primer binding site.J Virol. 1994 Mar;68(3):1605-14. doi: 10.1128/JVI.68.3.1605-1614.1994. J Virol. 1994. PMID: 7508999 Free PMC article.
-
Modulation of HIV-1 replication by RNA interference.Nature. 2002 Jul 25;418(6896):435-8. doi: 10.1038/nature00896. Epub 2002 Jun 26. Nature. 2002. PMID: 12087358 Free PMC article.
-
Inhibition of HIV-1 replication in vitro and in human infected cells by modified antisense oligonucleotides targeting the tRNALys3/RNA initiation complex.Antisense Nucleic Acid Drug Dev. 2001 Oct;11(5):301-15. doi: 10.1089/108729001753231687. Antisense Nucleic Acid Drug Dev. 2001. PMID: 11763347
-
The selective packaging and annealing of primer tRNALys3 in HIV-1.Curr HIV Res. 2004 Apr;2(2):163-75. doi: 10.2174/1570162043484988. Curr HIV Res. 2004. PMID: 15078180 Review.
-
Aspects of HIV-1 assembly that promote primer tRNA(Lys3) annealing to viral RNA.Virus Res. 2012 Nov;169(2):340-8. doi: 10.1016/j.virusres.2012.06.001. Epub 2012 Jun 12. Virus Res. 2012. PMID: 22698876 Review.
Cited by
-
Transfer RNA-derived fragment tRF-28-QSZ34KRQ590K in plasma exosomes may be a potential biomarker for atopic dermatitis in pediatric patients.Exp Ther Med. 2021 May;21(5):489. doi: 10.3892/etm.2021.9920. Epub 2021 Mar 16. Exp Ther Med. 2021. PMID: 33790998 Free PMC article.
-
HIV-1 transcription and latency: an update.Retrovirology. 2013 Jun 26;10:67. doi: 10.1186/1742-4690-10-67. Retrovirology. 2013. PMID: 23803414 Free PMC article. Review.
-
Effects of tRNA-derived fragments and microRNAs regulatory network on pancreatic acinar intracellular trypsinogen activation.Bioengineered. 2022 Feb;13(2):3207-3220. doi: 10.1080/21655979.2021.2018880. Bioengineered. 2022. PMID: 35045793 Free PMC article.
-
Tie-Break: Host and Retrotransposons Play tRNA.Trends Cell Biol. 2018 Oct;28(10):793-806. doi: 10.1016/j.tcb.2018.05.006. Epub 2018 Jun 19. Trends Cell Biol. 2018. PMID: 29934075 Free PMC article. Review.
-
The HIV Nef protein modulates cellular and exosomal miRNA profiles in human monocytic cells.J Extracell Vesicles. 2014 Mar 25;3. doi: 10.3402/jev.v3.23129. eCollection 2014. J Extracell Vesicles. 2014. PMID: 24678387 Free PMC article.
References
-
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. - PubMed
-
- Nelson P, Kiriakidou M, Sharma A, Maniataki E, Mourelatos Z. The microRNA world: small is mighty. Trends Biochem. Sci. 2003;28:534–540. - PubMed
-
- Gan J, Tropea JE, Austin BP, Court DL, Waugh DS, Ji X. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell. 2006;124:355–366. - PubMed
-
- Yeung ML, Bennasser Y, Le SY, Jeang KT. siRNA, miRNA and HIV: promises and challenges. Cell Res. 2005;15:935–946. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

