p53 responsive elements in human retrotransposons
- PMID: 19718052
- PMCID: PMC3193277
- DOI: 10.1038/onc.2009.246
p53 responsive elements in human retrotransposons
Abstract
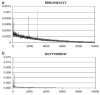
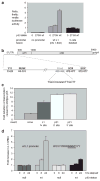
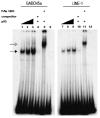
Long interspersed nuclear elements-1 (L1s) are highly repetitive DNA elements that are capable of altering the human genome through retrotransposition. To protect against L1 retroposition, the cell downregulates the expression of L1 proteins by various mechanisms, including high-density cytosine methylation of L1 promoters and DICER-dependent destruction of L1 mRNAs. In this report, a large number of p53 responsive elements, or p53 DNA binding sites, were detected in L1 elements within the human genome. At least some of these p53 responsive elements are functional and can act to increase the levels of L1 mRNA expression. The p53 protein can directly bind to a short 15-nucleotide sequence within the L1 promoter. This p53 responsive element within L1 is a recent addition to evolution, appearing approximately 20 million years ago. This suggests an interplay between L1 elements, which have a rich history of causing changes in the genome, and the p53 protein, the function of which is to protect against genomic changes. To understand these observations, a model is proposed in which the increased expression of L1 mRNAs by p53 actually increases, rather than decreases, the genomic stability through amplification of p53-dependent processes for genomic protection.
Conflict of interest statement
Figures



Similar articles
-
Immune signatures correlate with L1 retrotransposition in gastrointestinal cancers.Genome Res. 2018 Aug;28(8):1136-1146. doi: 10.1101/gr.231837.117. Epub 2018 Jul 3. Genome Res. 2018. PMID: 29970450 Free PMC article.
-
Control of chicken CR1 retrotransposons is independent of Dicer-mediated RNA interference pathway.BMC Biol. 2009 Aug 19;7:53. doi: 10.1186/1741-7007-7-53. BMC Biol. 2009. PMID: 19691826 Free PMC article.
-
Hypomethylation of intragenic LINE-1 represses transcription in cancer cells through AGO2.PLoS One. 2011 Mar 15;6(3):e17934. doi: 10.1371/journal.pone.0017934. PLoS One. 2011. PMID: 21423624 Free PMC article.
-
Control of LINE-1 Expression Maintains Genome Integrity in Germline and Early Embryo Development.Reprod Sci. 2022 Feb;29(2):328-340. doi: 10.1007/s43032-021-00461-1. Epub 2021 Jan 22. Reprod Sci. 2022. PMID: 33481218 Review.
-
LINE-1 retrotransposons: modulators of quantity and quality of mammalian gene expression?Bioessays. 2005 Aug;27(8):775-84. doi: 10.1002/bies.20257. Bioessays. 2005. PMID: 16015595 Review.
Cited by
-
Activating STING1-dependent immune signaling in TP53 mutant and wild-type acute myeloid leukemia.Proc Natl Acad Sci U S A. 2022 Jul 5;119(27):e2123227119. doi: 10.1073/pnas.2123227119. Epub 2022 Jun 27. Proc Natl Acad Sci U S A. 2022. PMID: 35759659 Free PMC article.
-
p53-Autophagy-Metastasis Link.Cancers (Basel). 2018 May 18;10(5):148. doi: 10.3390/cancers10050148. Cancers (Basel). 2018. PMID: 29783720 Free PMC article. Review.
-
How retrotransposons shape genome regulation.Curr Opin Genet Dev. 2016 Apr;37:90-100. doi: 10.1016/j.gde.2016.01.001. Epub 2016 Feb 6. Curr Opin Genet Dev. 2016. PMID: 26855260 Free PMC article. Review.
-
p53 binding sites in normal and cancer cells are characterized by distinct chromatin context.Cell Cycle. 2017;16(21):2073-2085. doi: 10.1080/15384101.2017.1361064. Epub 2017 Sep 28. Cell Cycle. 2017. PMID: 28820292 Free PMC article.
-
Restricting retrotransposons: a review.Mob DNA. 2016 Aug 11;7:16. doi: 10.1186/s13100-016-0070-z. eCollection 2016. Mob DNA. 2016. PMID: 27525044 Free PMC article. Review.
References
-
- Babushok DV, Kazazian HH., Jr Progress in understanding the biology of the human mutagen LINE-1. Hum Mutat. 2007;28:527–539. - PubMed
-
- Bensaad K, Vousden KH. Savior and slayer: the two faces of p53. Nat Med. 2005;11:1278–1279. - PubMed
-
- Budhram-Mahadeo V, Morris PJ, Smith MD, Midgley CA, Boxer LM, Latchman DS. p53 suppresses the activation of the Bcl-2 promoter by the Brn-3a POU family transcription factor. J Biol Chem. 1999;274:15237–15244. - PubMed
-
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

