ErbB2 resembles an autoinhibited invertebrate epidermal growth factor receptor
- PMID: 19718021
- PMCID: PMC2762480
- DOI: 10.1038/nature08297
ErbB2 resembles an autoinhibited invertebrate epidermal growth factor receptor
Abstract
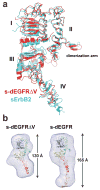
The orphan receptor tyrosine kinase ErbB2 (also known as HER2 or Neu) transforms cells when overexpressed, and it is an important therapeutic target in human cancer. Structural studies have suggested that the oncogenic (and ligand-independent) signalling properties of ErbB2 result from the absence of a key intramolecular 'tether' in the extracellular region that autoinhibits other human ErbB receptors, including the epidermal growth factor (EGF) receptor. Although ErbB2 is unique among the four human ErbB receptors, here we show that it is the closest structural relative of the single EGF receptor family member in Drosophila melanogaster (dEGFR). Genetic and biochemical data show that dEGFR is tightly regulated by growth factor ligands, yet a crystal structure shows that it, too, lacks the intramolecular tether seen in human EGFR, ErbB3 and ErbB4. Instead, a distinct set of autoinhibitory interdomain interactions hold unliganded dEGFR in an inactive state. All of these interactions are maintained (and even extended) in ErbB2, arguing against the suggestion that ErbB2 lacks autoinhibition. We therefore suggest that normal and pathogenic ErbB2 signalling may be regulated by ligands in the same way as dEGFR. Our findings have important implications for ErbB2 regulation in human cancer, and for developing therapeutic approaches that target novel aspects of this orphan receptor.
Conflict of interest statement
The authors declare that they have no competing financial interests.
Figures


Comment in
-
The ins and outs of EGFR asymmetry.Cell. 2010 Aug 20;142(4):513-5. doi: 10.1016/j.cell.2010.08.003. Cell. 2010. PMID: 20723751
Similar articles
-
A single ligand is sufficient to activate EGFR dimers.Proc Natl Acad Sci U S A. 2012 Jul 3;109(27):10861-6. doi: 10.1073/pnas.1201114109. Epub 2012 Jun 14. Proc Natl Acad Sci U S A. 2012. PMID: 22699492 Free PMC article.
-
Argos inhibits epidermal growth factor receptor signalling by ligand sequestration.Nature. 2004 Aug 26;430(7003):1040-4. doi: 10.1038/nature02840. Nature. 2004. PMID: 15329724
-
Structural basis for EGFR ligand sequestration by Argos.Nature. 2008 Jun 26;453(7199):1271-5. doi: 10.1038/nature06978. Epub 2008 May 25. Nature. 2008. PMID: 18500331 Free PMC article.
-
Small molecule inhibitors targeting the EGFR/ErbB family of protein-tyrosine kinases in human cancers.Pharmacol Res. 2019 Jan;139:395-411. doi: 10.1016/j.phrs.2018.11.014. Epub 2018 Nov 27. Pharmacol Res. 2019. PMID: 30500458 Review.
-
ErbB Receptors and Cancer.Methods Mol Biol. 2017;1652:3-35. doi: 10.1007/978-1-4939-7219-7_1. Methods Mol Biol. 2017. PMID: 28791631 Review.
Cited by
-
Affecting Rhomboid-3 function causes a dilated heart in adult Drosophila.PLoS Genet. 2010 May 27;6(5):e1000969. doi: 10.1371/journal.pgen.1000969. PLoS Genet. 2010. PMID: 20523889 Free PMC article.
-
Inhibition of ErbB3 by a monoclonal antibody that locks the extracellular domain in an inactive configuration.Proc Natl Acad Sci U S A. 2015 Oct 27;112(43):13225-30. doi: 10.1073/pnas.1518361112. Epub 2015 Oct 12. Proc Natl Acad Sci U S A. 2015. PMID: 26460020 Free PMC article.
-
Rigidity of the extracellular part of HER2: Evidence from engineering subdomain interfaces and shared-helix DARPin-DARPin fusions.Protein Sci. 2017 Sep;26(9):1796-1806. doi: 10.1002/pro.3216. Epub 2017 Jul 5. Protein Sci. 2017. PMID: 28639341 Free PMC article.
-
Mechanisms for kinase-mediated dimerization of the epidermal growth factor receptor.J Biol Chem. 2012 Nov 2;287(45):38244-53. doi: 10.1074/jbc.M112.414391. Epub 2012 Sep 17. J Biol Chem. 2012. PMID: 22988250 Free PMC article.
-
Structural basis for negative cooperativity in growth factor binding to an EGF receptor.Cell. 2010 Aug 20;142(4):568-79. doi: 10.1016/j.cell.2010.07.015. Cell. 2010. PMID: 20723758 Free PMC article.
References
-
- Di Fiore PP et al. erbB-2 is a potent oncogene when overexpressed in NIH/3T3 cells. Science. 1987;237:178–182. - PubMed
-
- Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–354. - PubMed
-
- Cho HS, et al. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003;421:756–760. - PubMed
-
- Garrett TP, et al. The crystal structure of a truncated ErbB2 ectodomain reveals an active conformation, poised to interact with other ErbB receptors. Mol Cell. 2003;11:495–505. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

