Congenital bovine spinal dysmyelination is caused by a missense mutation in the SPAST gene
- PMID: 19714378
- PMCID: PMC2854348
- DOI: 10.1007/s10048-009-0214-0
Congenital bovine spinal dysmyelination is caused by a missense mutation in the SPAST gene
Abstract
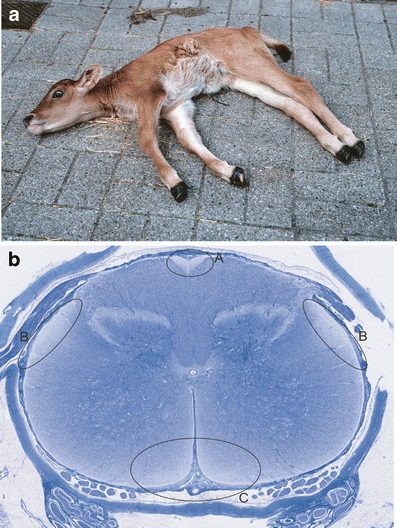
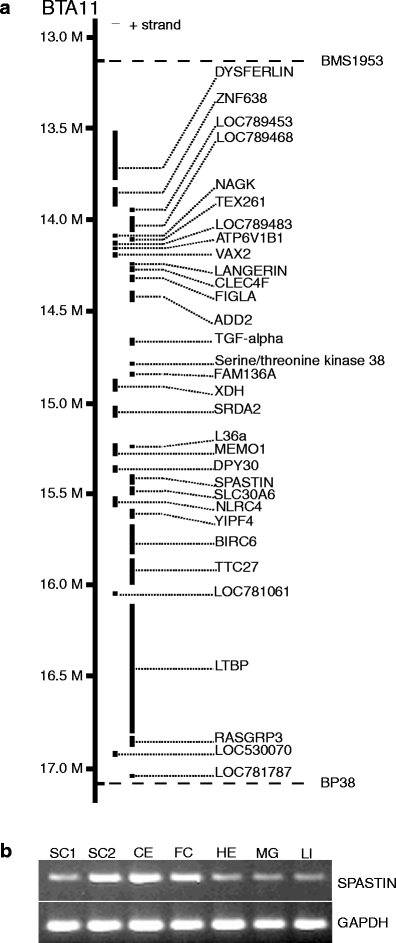
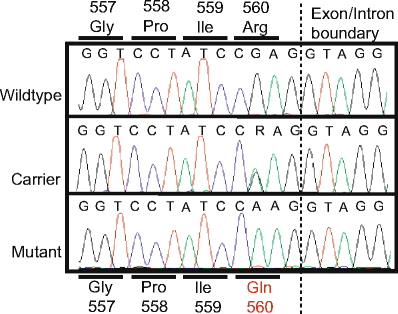
Bovine spinal dysmyelination (BSD) is a recessive congenital neurodegenerative disease in cattle (Bos taurus) characterized by pathological changes of the myelin sheaths in the spinal cord. The occurrence of BSD is a longstanding problem in the American Brown Swiss (ABS) breed and in several European cattle breeds upgraded with ABS. Here, we show that the disease locus on bovine chromosome 11 harbors the SPAST gene that, when mutated, is responsible for the human disorder hereditary spastic paraplegia (HSP). Initially, SPAST encoding Spastin was considered a less likely candidate gene for BSD since the modes of inheritance as well as the time of onset and severity of symptoms differ widely between HSP and BSD. However, sequence analysis of the bovine SPAST gene in affected animals identified a R560Q substitution at a position in the ATPase domain of the Spastin protein that is invariant from insects to mammals. Interestingly, three different mutations in human SPAST gene at the equivalent position are known to cause HSP. To explore this observation further, we genotyped more than 3,100 animals of various cattle breeds and found that the glutamine allele exclusively occurred in breeds upgraded with ABS. Furthermore, all confirmed BSD carriers were heterozygous, while all affected calves were homozygous for the glutamine allele consistent with recessive transmission of the underlying mutation and complete penetrance in the homozygous state. Subsequent analysis of recombinant Spastin in vitro showed that the R560Q substitution severely impaired the ATPase activity, demonstrating a causal relationship between the SPAST mutation and BSD.
Figures

Similar articles
-
SPAST mutations in Australian patients with hereditary spastic paraplegia.Intern Med J. 2012 Dec;42(12):1342-7. doi: 10.1111/j.1445-5994.2012.02941.x. Intern Med J. 2012. PMID: 23252998
-
Expansion of mutation spectrum, determination of mutation cluster regions and predictive structural classification of SPAST mutations in hereditary spastic paraplegia.Eur J Hum Genet. 2009 Feb;17(2):187-94. doi: 10.1038/ejhg.2008.147. Epub 2008 Aug 13. Eur J Hum Genet. 2009. PMID: 18701882 Free PMC article.
-
Spastin mutation screening in Chinese patients with pure hereditary spastic paraplegia.Parkinsonism Relat Disord. 2014 Aug;20(8):845-9. doi: 10.1016/j.parkreldis.2014.04.021. Epub 2014 May 2. Parkinsonism Relat Disord. 2014. PMID: 24824479
-
Detection of novel mutations and review of published data suggests that hereditary spastic paraplegia caused by spastin (SPAST) mutations is found more often in males.J Neurol Sci. 2011 Jul 15;306(1-2):62-5. doi: 10.1016/j.jns.2011.03.043. Epub 2011 May 5. J Neurol Sci. 2011. PMID: 21546041 Review.
-
Hereditary spastic paraplegia SPG4: what is known and not known about the disease.Brain. 2015 Sep;138(Pt 9):2471-84. doi: 10.1093/brain/awv178. Epub 2015 Jun 20. Brain. 2015. PMID: 26094131 Free PMC article. Review.
Cited by
-
Confirmation of a non-synonymous SNP in PNPLA8 as a candidate causal mutation for Weaver syndrome in Brown Swiss cattle.Genet Sel Evol. 2016 Mar 18;48:21. doi: 10.1186/s12711-016-0201-5. Genet Sel Evol. 2016. PMID: 26992691 Free PMC article.
-
Frameshift Variant in Novel Adenosine-A1-Receptor Homolog Associated With Bovine Spastic Syndrome/Late-Onset Bovine Spastic Paresis in Holstein Sires.Front Genet. 2020 Nov 30;11:591794. doi: 10.3389/fgene.2020.591794. eCollection 2020. Front Genet. 2020. PMID: 33329738 Free PMC article.
-
A missense mutation in TUBD1 is associated with high juvenile mortality in Braunvieh and Fleckvieh cattle.BMC Genomics. 2016 May 25;17:400. doi: 10.1186/s12864-016-2742-y. BMC Genomics. 2016. PMID: 27225349 Free PMC article.
-
Use of SNP genotypes to identify carriers of harmful recessive mutations in cattle populations.BMC Genomics. 2016 Nov 3;17(1):857. doi: 10.1186/s12864-016-3218-9. BMC Genomics. 2016. PMID: 27809787 Free PMC article.
-
An unusual splice defect in the mitofusin 2 gene (MFN2) is associated with degenerative axonopathy in Tyrolean Grey cattle.PLoS One. 2011 Apr 15;6(4):e18931. doi: 10.1371/journal.pone.0018931. PLoS One. 2011. PMID: 21526202 Free PMC article.
References
-
- Charlier C, Coppieters W, Rollin F, Desmecht D, Agerholm JS, Cambisano N, Carta E, Dardano S, Dive M, Fasquelle C, Frennet JC, Hanset R, Hubin X, Jorgensen C, Karim L, Kent M, Harvey K, Pearce BR, Simon P, Tama N, Nie H, Vandeputte S, Lien S, Longeri M, Fredholm M, Harvey RJ, Georges M. Highly effective SNP-based association mapping and management of recessive defects in livestock. Nat Genet. 2008;40:449–454. doi: 10.1038/ng.96. - DOI - PubMed
-
- Agerholm JS, Bendixen C, Andersen O, Arnbjerg J. Complex vertebral malformation in holstein calves. J Vet Diagn Invest. 2001;13:283–289. - PubMed
-
- Thomsen B, Horn P, Panitz F, Bendixen E, Petersen AH, Holm LE, Nielsen VH, Agerholm JS, Arnbjerg J, Bendixen C. A missense mutation in the bovine SLC35A3 gene, encoding a UDP-N-acetylglucosamine transporter, causes complex vertebral malformation. Genome Res. 2006;16:97–105. doi: 10.1101/gr.3690506. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

