Editing independent effects of ADARs on the miRNA/siRNA pathways
- PMID: 19713932
- PMCID: PMC2735678
- DOI: 10.1038/emboj.2009.244
Editing independent effects of ADARs on the miRNA/siRNA pathways
Abstract
Adenosine deaminases acting on RNA (ADARs) are best known for altering the coding sequences of mRNA through RNA editing, as in the GluR-B Q/R site. ADARs have also been shown to affect RNA interference (RNAi) and microRNA processing by deamination of specific adenosines to inosine. Here, we show that ADAR proteins can affect RNA processing independently of their enzymatic activity. We show that ADAR2 can modulate the processing of mir-376a2 independently of catalytic RNA editing activity. In addition, in a Drosophila assay for RNAi deaminase-inactive ADAR1 inhibits RNAi through the siRNA pathway. These results imply that ADAR1 and ADAR2 have biological functions as RNA-binding proteins that extend beyond editing per se and that even genomically encoded ADARs that are catalytically inactive may have such functions.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
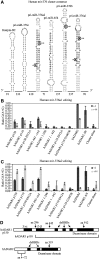
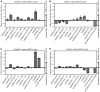
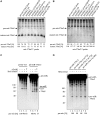

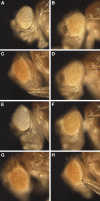
Similar articles
-
A comparative analysis of ADAR mutant mice reveals site-specific regulation of RNA editing.RNA. 2020 Apr;26(4):454-469. doi: 10.1261/rna.072728.119. Epub 2020 Jan 15. RNA. 2020. PMID: 31941663 Free PMC article.
-
Modulation of microRNA processing and expression through RNA editing by ADAR deaminases.Nat Struct Mol Biol. 2006 Jan;13(1):13-21. doi: 10.1038/nsmb1041. Epub 2005 Dec 20. Nat Struct Mol Biol. 2006. PMID: 16369484 Free PMC article.
-
Base-pairing probability in the microRNA stem region affects the binding and editing specificity of human A-to-I editing enzymes ADAR1-p110 and ADAR2.RNA Biol. 2018;15(7):976-989. doi: 10.1080/15476286.2018.1486658. Epub 2018 Jul 24. RNA Biol. 2018. PMID: 29950133 Free PMC article.
-
A-to-I editing of coding and non-coding RNAs by ADARs.Nat Rev Mol Cell Biol. 2016 Feb;17(2):83-96. doi: 10.1038/nrm.2015.4. Epub 2015 Dec 9. Nat Rev Mol Cell Biol. 2016. PMID: 26648264 Free PMC article. Review.
-
ADAR RNA editing in innate immune response phasing, in circadian clocks and in sleep.Biochim Biophys Acta Gene Regul Mech. 2019 Mar;1862(3):356-369. doi: 10.1016/j.bbagrm.2018.10.011. Epub 2018 Oct 31. Biochim Biophys Acta Gene Regul Mech. 2019. PMID: 30391332 Review.
Cited by
-
Reciprocal regulation of A-to-I RNA editing and the vertebrate nervous system.Front Neurosci. 2013 Apr 18;7:61. doi: 10.3389/fnins.2013.00061. eCollection 2013. Front Neurosci. 2013. PMID: 23616744 Free PMC article.
-
Defective DROSHA processing contributes to downregulation of MiR-15/-16 in chronic lymphocytic leukemia.Leukemia. 2014 Jan;28(1):98-107. doi: 10.1038/leu.2013.246. Epub 2013 Aug 26. Leukemia. 2014. PMID: 23974981
-
ADAR proteins: double-stranded RNA and Z-DNA binding domains.Curr Top Microbiol Immunol. 2012;353:35-60. doi: 10.1007/82_2011_145. Curr Top Microbiol Immunol. 2012. PMID: 21728134 Free PMC article. Review.
-
Emerging role of the RNA-editing enzyme ADAR1 in stem cell fate and function.Biomark Res. 2023 Jun 6;11(1):61. doi: 10.1186/s40364-023-00503-7. Biomark Res. 2023. PMID: 37280687 Free PMC article. Review.
-
Adar-mediated A-to-I editing is required for embryonic patterning and innate immune response regulation in zebrafish.Nat Commun. 2022 Sep 20;13(1):5520. doi: 10.1038/s41467-022-33260-6. Nat Commun. 2022. PMID: 36127363 Free PMC article.
References
-
- Beghini A, Ripamonti CB, Peterlongo P, Roversi G, Cairoli R, Morra E, Larizza L (2000) RNA hyperediting and alternative splicing of hematopoietic cell phosphatase (PTPN6) gene in acute myeloid leukemia. Hum Mol Genet 9: 2297–2304 - PubMed
-
- Cenci C, Barzotti R, Galeano F, Corbelli S, Rota R, Massimi L, Di Rocco C, O'Connell MA, Gallo A (2008) Down-regulation of RNA editing in pediatric astrocytomas: ADAR2 editing activity inhibits cell migration and proliferation. J Biol Chem 283: 7251–7260 - PubMed
-
- Connolly CM, Dearth AT, Braun RE (2005) Disruption of murine Tenr results in teratospermia and male infertility. Dev Biol 278: 13–21 - PubMed
-
- Desterro JM, Keegan LP, Lafarga M, Berciano MT, O'Connell M, Carmo-Fonseca M (2003) Dynamic association of RNA-editing enzymes with the nucleolus. J Cell Sci 116: 1805–1818 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

