Weighted gene co-expression network analysis of the peripheral blood from Amyotrophic Lateral Sclerosis patients
- PMID: 19712483
- PMCID: PMC2743717
- DOI: 10.1186/1471-2164-10-405
Weighted gene co-expression network analysis of the peripheral blood from Amyotrophic Lateral Sclerosis patients
Abstract
Background: Amyotrophic Lateral Sclerosis (ALS) is a lethal disorder characterized by progressive degeneration of motor neurons in the brain and spinal cord. Diagnosis is mainly based on clinical symptoms, and there is currently no therapy to stop the disease or slow its progression. Since access to spinal cord tissue is not possible at disease onset, we investigated changes in gene expression profiles in whole blood of ALS patients.
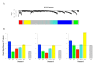
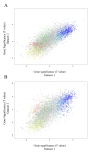
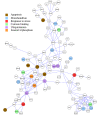
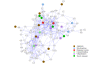
Results: Our transcriptional study showed dramatic changes in blood of ALS patients; 2,300 probes (9.4%) showed significant differential expression in a discovery dataset consisting of 30 ALS patients and 30 healthy controls. Weighted gene co-expression network analysis (WGCNA) was used to find disease-related networks (modules) and disease related hub genes. Two large co-expression modules were found to be associated with ALS. Our findings were replicated in a second (30 patients and 30 controls) and third dataset (63 patients and 63 controls), thereby demonstrating a highly significant and consistent association of two large co-expression modules with ALS disease status. Ingenuity Pathway Analysis of the ALS related module genes implicates enrichment of functional categories related to genetic disorders, neurodegeneration of the nervous system and inflammatory disease. The ALS related modules contain a number of candidate genes possibly involved in pathogenesis of ALS.
Conclusion: This first large-scale blood gene expression study in ALS observed distinct patterns between cases and controls which may provide opportunities for biomarker development as well as new insights into the molecular mechanisms of the disease.
Figures
Similar articles
-
Microarray analysis of peripheral blood lymphocytes from ALS patients and the SAFE detection of the KEGG ALS pathway.BMC Med Genomics. 2011 Oct 25;4:74. doi: 10.1186/1755-8794-4-74. BMC Med Genomics. 2011. PMID: 22027401 Free PMC article.
-
Identification of potentially functional modules and diagnostic genes related to amyotrophic lateral sclerosis based on the WGCNA and LASSO algorithms.Sci Rep. 2022 Nov 22;12(1):20144. doi: 10.1038/s41598-022-24306-2. Sci Rep. 2022. PMID: 36418457 Free PMC article.
-
RNAseq Analyses Identify Tumor Necrosis Factor-Mediated Inflammation as a Major Abnormality in ALS Spinal Cord.PLoS One. 2016 Aug 3;11(8):e0160520. doi: 10.1371/journal.pone.0160520. eCollection 2016. PLoS One. 2016. PMID: 27487029 Free PMC article.
-
New insights into the gene expression associated to amyotrophic lateral sclerosis.Life Sci. 2018 Jan 15;193:110-123. doi: 10.1016/j.lfs.2017.12.016. Epub 2017 Dec 11. Life Sci. 2018. PMID: 29241710 Review.
-
Transcriptomics in amyotrophic lateral sclerosis.Front Biosci (Elite Ed). 2018 Jan 1;10(1):103-121. doi: 10.2741/e811. Front Biosci (Elite Ed). 2018. PMID: 28930607 Review.
Cited by
-
Systems analysis of human brain gene expression: mechanisms for HIV-associated neurocognitive impairment and common pathways with Alzheimer's disease.BMC Med Genomics. 2013 Feb 13;6:4. doi: 10.1186/1755-8794-6-4. BMC Med Genomics. 2013. PMID: 23406646 Free PMC article.
-
Biological networks and complexity in early-onset motor neuron diseases.Front Neurol. 2022 Oct 21;13:1035406. doi: 10.3389/fneur.2022.1035406. eCollection 2022. Front Neurol. 2022. PMID: 36341099 Free PMC article.
-
SMA-MAP: a plasma protein panel for spinal muscular atrophy.PLoS One. 2013;8(4):e60113. doi: 10.1371/journal.pone.0060113. Epub 2013 Apr 2. PLoS One. 2013. PMID: 23565191 Free PMC article.
-
Common variant at 16p11.2 conferring risk of psychosis.Mol Psychiatry. 2014 Jan;19(1):108-14. doi: 10.1038/mp.2012.157. Epub 2012 Nov 20. Mol Psychiatry. 2014. PMID: 23164818 Free PMC article.
-
Microarray analysis of peripheral blood lymphocytes from ALS patients and the SAFE detection of the KEGG ALS pathway.BMC Med Genomics. 2011 Oct 25;4:74. doi: 10.1186/1755-8794-4-74. BMC Med Genomics. 2011. PMID: 22027401 Free PMC article.
References
-
- Schymick JC, Scholz SW, Fung HC, Britton A, Arepalli S, Gibbs JR, Lombardo F, Matarin M, Kasperaviciute D, Hernandez DG, et al. Genome-wide genotyping in amyotrophic lateral sclerosis and neurologically normal controls: first stage analysis and public release of data. Lancet Neurol. 2007;6:322–328. doi: 10.1016/S1474-4422(07)70037-6. - DOI - PubMed
-
- van Es MA, Van Vught PW, Blauw HM, Franke L, Saris CG, Andersen PM, Bosch L Van Den, de Jong SW, van 't Slot R, Birve A, et al. ITPR2 as a susceptibility gene in sporadic amyotrophic lateral sclerosis: a genome-wide association study. Lancet Neurol. 2007;6:869–877. doi: 10.1016/S1474-4422(07)70222-3. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

