Protein phosphatase 2A-dependent dephosphorylation of replication protein A is required for the repair of DNA breaks induced by replication stress
- PMID: 19704001
- PMCID: PMC2772729
- DOI: 10.1128/MCB.00191-09
Protein phosphatase 2A-dependent dephosphorylation of replication protein A is required for the repair of DNA breaks induced by replication stress
Abstract
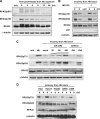
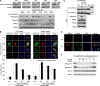
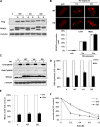
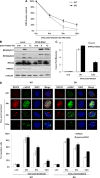
Eukaryotic genomic integrity is safeguarded by cell cycle checkpoints and DNA repair pathways, collectively known as the DNA damage response, wherein replication protein A (RPA) is a key regulator playing multiple critical roles. The genotoxic insult-induced phosphorylation of the 32-kDa subunit of human RPA (RPA32), most notably the ATM/ATR-dependent phosphorylation at T21 and S33, acts to suppress DNA replication and recruit other checkpoint/repair proteins to the DNA lesions. It is not clear, however, how the DNA damage-responsive function of phosphorylated RPA is attenuated and how the replication-associated activity of the unphosphorylated form of RPA is restored when cells start to resume the normal cell cycle. We report here that in cells recovering from hydroxyurea (HU)-induced genotoxic stress, RPA32 is dephosphorylated by the serine/threonine protein phosphatase 2A (PP2A). Interference with PP2A catalytic activity causes persistent RPA32 phosphorylation and increased HU sensitivity. The PP2A catalytic subunit binds to RPA following DNA damage and can dephosphorylate RPA32 in vitro. Cells expressing a RPA32 persistent phosphorylation mimetic exhibit normal checkpoint activation and reenter the cell cycle normally after recovery but display a pronounced defect in the repair of DNA breaks. These data indicate that PP2A-mediated RPA32 dephosphorylation is required for the efficient DNA damage repair.
Figures

Similar articles
-
Distinct roles for DNA-PK, ATM and ATR in RPA phosphorylation and checkpoint activation in response to replication stress.Nucleic Acids Res. 2012 Nov;40(21):10780-94. doi: 10.1093/nar/gks849. Epub 2012 Sep 12. Nucleic Acids Res. 2012. PMID: 22977173 Free PMC article.
-
Phosphatidyl inositol 3-kinase-like serine/threonine protein kinases (PIKKs) are required for DNA damage-induced phosphorylation of the 32 kDa subunit of replication protein A at threonine 21.Nucleic Acids Res. 2004 Feb 10;32(3):997-1005. doi: 10.1093/nar/gkh265. Print 2004. Nucleic Acids Res. 2004. PMID: 14872059 Free PMC article.
-
Phosphorylation of the replication protein A large subunit in the Saccharomyces cerevisiae checkpoint response.Nucleic Acids Res. 2000 Oct 1;28(19):3725-32. doi: 10.1093/nar/28.19.3725. Nucleic Acids Res. 2000. PMID: 11000264 Free PMC article.
-
Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses.J Cell Physiol. 2006 Aug;208(2):267-73. doi: 10.1002/jcp.20622. J Cell Physiol. 2006. PMID: 16523492 Free PMC article. Review.
-
Protein kinases that regulate chromosome stability and their downstream targets.Genome Dyn. 2006;1:131-148. doi: 10.1159/000092505. Genome Dyn. 2006. PMID: 18724058 Review.
Cited by
-
DNA Damage Response in Early Breast Cancer: A Phase III Cohort in the Phobos Study.Cancers (Basel). 2024 Jul 23;16(15):2628. doi: 10.3390/cancers16152628. Cancers (Basel). 2024. PMID: 39123356 Free PMC article.
-
Phosphatases in the cellular response to DNA damage.Cell Commun Signal. 2010 Sep 22;8:27. doi: 10.1186/1478-811X-8-27. Cell Commun Signal. 2010. PMID: 20860841 Free PMC article.
-
Rfa2 is specifically dephosphorylated by Pph3 in Candida albicans.Biochem J. 2013 Feb 1;449(3):673-81. doi: 10.1042/BJ20120952. Biochem J. 2013. PMID: 23140133 Free PMC article.
-
PP2A is activated by cytochrome c upon formation of a diffuse encounter complex with SET/TAF-Iβ.Comput Struct Biotechnol J. 2022 Jul 8;20:3695-3707. doi: 10.1016/j.csbj.2022.07.009. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35891793 Free PMC article.
-
Cell Cycle and DNA Repair Regulation in the Damage Response: Protein Phosphatases Take Over the Reins.Int J Mol Sci. 2020 Jan 10;21(2):446. doi: 10.3390/ijms21020446. Int J Mol Sci. 2020. PMID: 31936707 Free PMC article. Review.
References
-
- Abraham, R. T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15:2177-2196. - PubMed
-
- Bartek, J., and J. Lukas. 2007. DNA damage checkpoints: from initiation to recovery or adaptation. Curr. Opin. Cell Biol. 19:238-245. - PubMed
-
- Binz, S. K., Y. Lao, D. F. Lowry, and M. S. Wold. 2003. The phosphorylation domain of the 32-kDa subunit of replication protein A (RPA) modulates RPA-DNA interactions. Evidence for an intersubunit interaction. J. Biol. Chem. 278:35584-35591. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
