Chemoattractant signaling between tumor cells and macrophages regulates cancer cell migration, metastasis and neovascularization
- PMID: 19696929
- PMCID: PMC2725301
- DOI: 10.1371/journal.pone.0006713
Chemoattractant signaling between tumor cells and macrophages regulates cancer cell migration, metastasis and neovascularization
Abstract
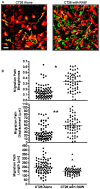
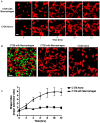
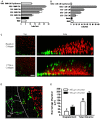
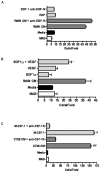
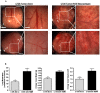
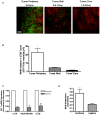
Tumor-associated macrophages are known to influence cancer progression by modulation of immune function, angiogenesis, and cell metastasis, however, little is known about the chemokine signaling networks that regulate this process. Utilizing CT26 colon cancer cells and RAW 264.7 macrophages as a model cellular system, we demonstrate that treatment of CT26 cells with RAW 264.7 conditioned medium induces cell migration, invasion and metastasis. Inflammatory gene microarray analysis indicated CT26-stimulated RAW 264.7 macrophages upregulate SDF-1alpha and VEGF, and that these cytokines contribute to CT26 migration in vitro. RAW 264.7 macrophages also showed a robust chemotactic response towards CT26-derived chemokines. In particular, microarray analysis and functional testing revealed CSF-1 as the major chemoattractant for RAW 264.7 macrophages. Interestingly, in the chick CAM model of cancer progression, RAW 264.7 macrophages localized specifically to the tumor periphery where they were found to increase CT26 tumor growth, microvascular density, vascular disruption, and lung metastasis, suggesting these cells home to actively invading areas of the tumor, but not the hypoxic core of the tumor mass. In support of these findings, hypoxic conditions down regulated CSF-1 production in several tumor cell lines and decreased RAW 264.7 macrophage migration in vitro. Together our findings suggest a model where normoxic tumor cells release CSF-1 to recruit macrophages to the tumor periphery where they secrete motility and angiogenic factors that facilitate tumor cell invasion and metastasis.
Conflict of interest statement
Figures

Similar articles
-
Stromal cell-derived factor-1 promotes cell migration and tumor growth of colorectal metastasis.Neoplasia. 2007 Oct;9(10):862-70. doi: 10.1593/neo.07559. Neoplasia. 2007. PMID: 17971906 Free PMC article.
-
Chemoattraction of macrophages by secretory molecules derived from cells expressing the signal peptide of eosinophil cationic protein.BMC Syst Biol. 2012 Aug 20;6:105. doi: 10.1186/1752-0509-6-105. BMC Syst Biol. 2012. PMID: 22906315 Free PMC article.
-
Paracrine regulation of vascular endothelial growth factor--a expression during macrophage-melanoma cell interaction: role of monocyte chemotactic protein-1 and macrophage colony-stimulating factor.J Interferon Cytokine Res. 2005 Nov;25(11):674-83. doi: 10.1089/jir.2005.25.674. J Interferon Cytokine Res. 2005. PMID: 16318581
-
Monocytes and dendritic cells in a hypoxic environment: Spotlights on chemotaxis and migration.Immunobiology. 2008;213(9-10):733-49. doi: 10.1016/j.imbio.2008.07.031. Epub 2008 Sep 21. Immunobiology. 2008. PMID: 18926289 Review.
-
Distinct role of macrophages in different tumor microenvironments.Cancer Res. 2006 Jan 15;66(2):605-12. doi: 10.1158/0008-5472.CAN-05-4005. Cancer Res. 2006. PMID: 16423985 Review.
Cited by
-
Tumor angiogenesis: MMP-mediated induction of intravasation- and metastasis-sustaining neovasculature.Matrix Biol. 2015 May-Jul;44-46:94-112. doi: 10.1016/j.matbio.2015.04.004. Epub 2015 Apr 22. Matrix Biol. 2015. PMID: 25912949 Free PMC article. Review.
-
Intratumoral macrophages contribute to epithelial-mesenchymal transition in solid tumors.BMC Cancer. 2012 Jan 24;12:35. doi: 10.1186/1471-2407-12-35. BMC Cancer. 2012. PMID: 22273460 Free PMC article.
-
Differential Oxygenation in Tumor Microenvironment Modulates Macrophage and Cancer Cell Crosstalk: Novel Experimental Setting and Proof of Concept.Front Oncol. 2019 Feb 6;9:43. doi: 10.3389/fonc.2019.00043. eCollection 2019. Front Oncol. 2019. PMID: 30788287 Free PMC article.
-
Antibodies for the Treatment of Brain Metastases, a Dream or a Reality?Pharmaceutics. 2020 Jan 13;12(1):62. doi: 10.3390/pharmaceutics12010062. Pharmaceutics. 2020. PMID: 31940974 Free PMC article. Review.
-
Macrophage-tumor crosstalk: role of TAMR tyrosine kinase receptors and of their ligands.Cell Mol Life Sci. 2012 May;69(9):1391-414. doi: 10.1007/s00018-011-0863-7. Epub 2011 Nov 11. Cell Mol Life Sci. 2012. PMID: 22076650 Free PMC article. Review.
References
-
- Tlsty TD, Coussens LM. Tumor stroma and regulation of cancer development. Annu Rev Pathol. 2006;1:119–150. - PubMed
-
- Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat Rev Cancer. 2004;4:71–78. - PubMed
-
- Sica A, Allavena P, Mantovani A. Cancer related inflammation: the macrophage connection. Cancer Lett. 2008;267:204–215. - PubMed
-
- Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23:549–555. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

