Sensitive digital quantification of DNA methylation in clinical samples
- PMID: 19684580
- PMCID: PMC2847606
- DOI: 10.1038/nbt.1559
Sensitive digital quantification of DNA methylation in clinical samples
Abstract
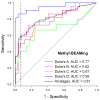
Analysis of abnormally methylated genes is increasingly important in basic research and in the development of cancer biomarkers. We have developed methyl-BEAMing technology to enable absolute quantification of the number of methylated molecules in a sample. Individual DNA fragments are amplified and analyzed either by flow cytometry or next-generation sequencing. We demonstrate enumeration of as few as one methylated molecule in approximately 5,000 unmethylated molecules in DNA from plasma or fecal samples. Using methylated vimentin as a biomarker in plasma samples, methyl-BEAMing detected 59% of cancer cases. In early-stage colorectal cancers, this sensitivity was four times more than that obtained by assaying serum-carcinoembryonic antigen (CEA). With stool samples, methyl-BEAMing detected 41% of cancers and 45% of advanced adenomas. In addition to diagnostic and prognostic applications, this digital quantification of rare methylation events should be applicable to preclinical assessment of new epigenetic biomarkers and quantitative analyses in epigenetic research.
Conflict of interest statement
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at
Figures

Similar articles
-
Stool methylation-specific polymerase chain reaction assay for the detection of colorectal neoplasia in Korean patients.Dis Colon Rectum. 2009 Aug;52(8):1452-9; discussion 1459-63. doi: 10.1007/DCR.0b013e3181a79533. Dis Colon Rectum. 2009. PMID: 19617759
-
Aberrantly methylated CDKN2A, MGMT, and MLH1 in colon polyps and in fecal DNA from patients with colorectal polyps.Clin Cancer Res. 2005 Feb 1;11(3):1203-9. Clin Cancer Res. 2005. PMID: 15709190
-
A novel method to capture methylated human DNA from stool: implications for colorectal cancer screening.Clin Chem. 2007 Sep;53(9):1646-51. doi: 10.1373/clinchem.2007.086223. Clin Chem. 2007. PMID: 17712002
-
Stool DNA methylation assays in colorectal cancer screening.World J Gastroenterol. 2015 Sep 21;21(35):10057-61. doi: 10.3748/wjg.v21.i35.10057. World J Gastroenterol. 2015. PMID: 26401070 Free PMC article. Review.
-
DNA methylation patterns as noninvasive biomarkers and targets of epigenetic therapies in colorectal cancer.Epigenomics. 2016 May;8(5):685-703. doi: 10.2217/epi-2015-0013. Epub 2016 Apr 22. Epigenomics. 2016. PMID: 27102979 Free PMC article. Review.
Cited by
-
[Chances and risks of blood-based molecular pathological analysis of circulating tumor cells (CTC) and cell-free DNA (cfDNA) in personalized cancer therapy: positional paper from the study group on liquid biopsy of the working group for molecular pathology in the German Society of Pathology (DGP)].Pathologe. 2015 Feb;36(1):92-7. doi: 10.1007/s00292-014-2069-x. Pathologe. 2015. PMID: 25533324 Review. German. No abstract available.
-
Clinical Applications of Liquid Biopsies in Gastrointestinal Oncology.Gastrointest Cancer Res. 2014 Sep;7(4 Suppl 1):S8-S12. Gastrointest Cancer Res. 2014. PMID: 27053977 Free PMC article.
-
Artifactual mutations resulting from DNA lesions limit detection levels in ultrasensitive sequencing applications.DNA Res. 2016 Dec;23(6):547-559. doi: 10.1093/dnares/dsw038. Epub 2016 Jul 31. DNA Res. 2016. PMID: 27477585 Free PMC article.
-
Transcription factors as readers and effectors of DNA methylation.Nat Rev Genet. 2016 Aug 1;17(9):551-65. doi: 10.1038/nrg.2016.83. Nat Rev Genet. 2016. PMID: 27479905 Free PMC article. Review.
-
Epigenetics advancing personalized nanomedicine in cancer therapy.Adv Drug Deliv Rev. 2012 Oct;64(13):1532-43. doi: 10.1016/j.addr.2012.08.004. Epub 2012 Aug 19. Adv Drug Deliv Rev. 2012. PMID: 22921595 Free PMC article. Review.
References
-
- Sidransky D. Emerging molecular markers of cancer. Nat Rev Cancer. 2002;2:210–219. - PubMed
-
- Fleischhacker M, Schmidt B. Circulating nucleic acids (CNAs) and cancer--a survey. Biochim Biophys Acta. 2007;1775:181–232. - PubMed
-
- Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. - PubMed
-
- Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–266. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

