SOSS complexes participate in the maintenance of genomic stability
- PMID: 19683501
- PMCID: PMC2756616
- DOI: 10.1016/j.molcel.2009.06.011
SOSS complexes participate in the maintenance of genomic stability
Abstract
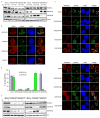
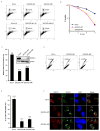
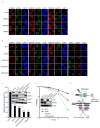
Proteins that bind to single-stranded DNA (ssDNA) are essential for DNA replication, recombinational repair, and maintenance of genomic stability. Here, we describe the characterization of an ssDNA-binding heterotrimeric complex, SOSS (sensor of ssDNA) in human, which consists of human SSB homologs hSSB1/2 (SOSS-B1/2) and INTS3 (SOSS-A) and a previously uncharacterized protein C9orf80 (SOSS-C). We have shown that SOSS-A serves as a central adaptor required not only for SOSS complex assembly and stability, but also for facilitating the accumulation of SOSS complex to DNA ends. Moreover, SOSS-depleted cells display increased ionizing radiation sensitivity, defective G2/M checkpoint, and impaired homologous recombination repair. Thus, our study defines a pathway involving the sensing of ssDNA by SOSS complex and suggests that this SOSS complex is likely involved in the maintenance of genome stability.
Figures

Comment in
-
SOSS1/2: Sensors of single-stranded DNA at a break.Mol Cell. 2009 Aug 14;35(3):258-9. doi: 10.1016/j.molcel.2009.07.016. Mol Cell. 2009. PMID: 19683490 Free PMC article.
Similar articles
-
SOSS1/2: Sensors of single-stranded DNA at a break.Mol Cell. 2009 Aug 14;35(3):258-9. doi: 10.1016/j.molcel.2009.07.016. Mol Cell. 2009. PMID: 19683490 Free PMC article.
-
The molecular details of a novel phosphorylation-dependent interaction between MRN and the SOSS complex.Protein Sci. 2023 Oct;32(10):e4782. doi: 10.1002/pro.4782. Protein Sci. 2023. PMID: 37705456 Free PMC article.
-
Biochemical characterization of INTS3 and C9ORF80, two subunits of hNABP1/2 heterotrimeric complex in nucleic acid binding.Biochem J. 2018 Jan 2;475(1):45-60. doi: 10.1042/BCJ20170351. Biochem J. 2018. PMID: 29150435 Free PMC article.
-
Human single-stranded DNA binding proteins: guardians of genome stability.Acta Biochim Biophys Sin (Shanghai). 2016 Jul;48(7):671-7. doi: 10.1093/abbs/gmw044. Epub 2016 May 23. Acta Biochim Biophys Sin (Shanghai). 2016. PMID: 27217471 Review.
-
[Regulation of homologous recombination: old and new roles of cyclin-dependent kinases].Tanpakushitsu Kakusan Koso. 2009 Mar;54(4 Suppl):479-84. Tanpakushitsu Kakusan Koso. 2009. PMID: 21089495 Review. Japanese. No abstract available.
Cited by
-
Mutant Presenilin 1 Dysregulates Exosomal Proteome Cargo Produced by Human-Induced Pluripotent Stem Cell Neurons.ACS Omega. 2021 May 13;6(20):13033-13056. doi: 10.1021/acsomega.1c00660. eCollection 2021 May 25. ACS Omega. 2021. PMID: 34056454 Free PMC article.
-
Human DNA2 possesses a cryptic DNA unwinding activity that functionally integrates with BLM or WRN helicases.Elife. 2016 Sep 9;5:e18574. doi: 10.7554/eLife.18574. Elife. 2016. PMID: 27612385 Free PMC article.
-
hSSB1 (NABP2/OBFC2B) modulates the DNA damage and androgen-induced transcriptional response in prostate cancer.Prostate. 2023 May;83(7):628-640. doi: 10.1002/pros.24496. Epub 2023 Feb 22. Prostate. 2023. PMID: 36811381 Free PMC article.
-
An integrated approach identifies new oncotargets in melanoma.Oncotarget. 2017 Dec 15;9(14):11489-11502. doi: 10.18632/oncotarget.23727. eCollection 2018 Feb 20. Oncotarget. 2017. PMID: 29545914 Free PMC article.
-
Integrator complex regulates NELF-mediated RNA polymerase II pause/release and processivity at coding genes.Nat Commun. 2014 Nov 20;5:5531. doi: 10.1038/ncomms6531. Nat Commun. 2014. PMID: 25410209 Free PMC article.
References
-
- Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–276. - PubMed
-
- Bartek J, Lukas C, Lukas J. Checking on DNA damage in S phase. Nat Rev Mol Cell Biol. 2004;5:792–804. - PubMed
-
- Bartek J, Lukas J. DNA damage checkpoints: from initiation to recovery or adaptation. Curr Opin Cell Biol. 2007;19:238–245. - PubMed
-
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. - PubMed
-
- Borde V. The multiple roles of the Mre11 complex for meiotic recombination. Chromosome Res. 2007;15:551–563. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

