A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity
- PMID: 19668189
- PMCID: PMC3624089
- DOI: 10.1038/nature08287
A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity
Abstract
The reprogramming of differentiated cells to pluripotent cells (induced pluripotent stem (iPS) cells) is known to be an inefficient process. We recently reported that cells with short telomeres cannot be reprogrammed to iPS cells despite their normal proliferation rates, probably reflecting the existence of 'reprogramming barriers' that abort the reprogramming of cells with uncapped telomeres. Here we show that p53 (also known as Trp53 in mice and TP53 in humans) is critically involved in preventing the reprogramming of cells carrying various types of DNA damage, including short telomeres, DNA repair deficiencies, or exogenously inflicted DNA damage. Reprogramming in the presence of pre-existing, but tolerated, DNA damage is aborted by the activation of a DNA damage response and p53-dependent apoptosis. Abrogation of p53 allows efficient reprogramming in the face of DNA damage and the generation of iPS cells carrying persistent DNA damage and chromosomal aberrations. These observations indicate that during reprogramming cells increase their intolerance to different types of DNA damage and that p53 is critical in preventing the generation of human and mouse pluripotent cells from suboptimal parental cells.
Figures
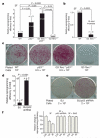
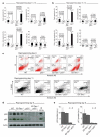
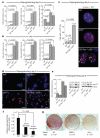
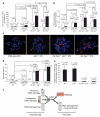
Comment in
-
Stem cells: The promises and perils of p53.Nature. 2009 Aug 27;460(7259):1085-6. doi: 10.1038/4601085a. Nature. 2009. PMID: 19713919 Free PMC article.
Similar articles
-
Linking the p53 tumour suppressor pathway to somatic cell reprogramming.Nature. 2009 Aug 27;460(7259):1140-4. doi: 10.1038/nature08311. Epub 2009 Aug 9. Nature. 2009. PMID: 19668186 Free PMC article.
-
Immortalization eliminates a roadblock during cellular reprogramming into iPS cells.Nature. 2009 Aug 27;460(7259):1145-8. doi: 10.1038/nature08285. Epub 2009 Aug 9. Nature. 2009. PMID: 19668190 Free PMC article.
-
p53 isoform Δ133p53 promotes efficiency of induced pluripotent stem cells and ensures genomic integrity during reprogramming.Sci Rep. 2016 Nov 22;6:37281. doi: 10.1038/srep37281. Sci Rep. 2016. PMID: 27874035 Free PMC article.
-
SOX2 and p53 Expression Control Converges in PI3K/AKT Signaling with Versatile Implications for Stemness and Cancer.Int J Mol Sci. 2020 Jul 11;21(14):4902. doi: 10.3390/ijms21144902. Int J Mol Sci. 2020. PMID: 32664542 Free PMC article. Review.
-
The role of p53-mediated apoptosis as a crucial anti-tumor response to genomic instability: lessons from mouse models.Mutat Res. 2005 Jan 6;569(1-2):145-57. doi: 10.1016/j.mrfmmm.2004.04.019. Mutat Res. 2005. PMID: 15603759 Review.
Cited by
-
A continuous molecular roadmap to iPSC reprogramming through progression analysis of single-cell mass cytometry.Cell Stem Cell. 2015 Mar 5;16(3):323-37. doi: 10.1016/j.stem.2015.01.015. Cell Stem Cell. 2015. PMID: 25748935 Free PMC article.
-
Loss of p53 exacerbates multiple myeloma phenotype by facilitating the reprogramming of hematopoietic stem/progenitor cells to malignant plasma cells by MafB.Cell Cycle. 2012 Oct 15;11(20):3896-900. doi: 10.4161/cc.22186. Epub 2012 Sep 14. Cell Cycle. 2012. PMID: 22983007 Free PMC article.
-
Direct lineage conversion: induced neuronal cells and induced neural stem cells.Protein Cell. 2012 Nov;3(11):826-33. doi: 10.1007/s13238-012-2068-8. Epub 2012 Sep 21. Protein Cell. 2012. PMID: 22996174 Free PMC article. Review.
-
Therapeutic targeting of the p53 pathway in cancer stem cells.Expert Opin Ther Targets. 2012 Dec;16(12):1161-74. doi: 10.1517/14728222.2012.726985. Epub 2012 Sep 24. Expert Opin Ther Targets. 2012. PMID: 22998602 Free PMC article. Review.
-
Genetic instability of modified stem cells - a first step towards malignant transformation?Am J Stem Cells. 2013 Mar 8;2(1):39-51. Print 2013. Am J Stem Cells. 2013. PMID: 23671815 Free PMC article.
References
-
- Marion RM, et al. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell. 2009;4:141–154. - PubMed
-
- Davy P, Allsopp R. Balancing out the ends during iPSC nuclear reprogramming. Cell Stem Cell. 2009;4:95–96. - PubMed
-
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. - PubMed
-
- Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

