Allosteric modulation of DNA by small molecules
- PMID: 19666554
- PMCID: PMC2726366
- DOI: 10.1073/pnas.0906532106
Allosteric modulation of DNA by small molecules
Abstract
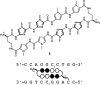
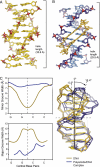
Many human diseases are caused by dysregulated gene expression. The oversupply of transcription factors may be required for the growth and metastatic behavior of human cancers. Cell permeable small molecules that can be programmed to disrupt transcription factor-DNA interfaces could silence aberrant gene expression pathways. Pyrrole-imidazole polyamides are DNA minor-groove binding molecules that are programmable for a large repertoire of DNA motifs. A high resolution X-ray crystal structure of an 8-ring cyclic Py/Im polyamide bound to the central 6 bp of the sequence d(5'-CCAGGCCTGG-3')2 reveals a 4 A widening of the minor groove and compression of the major groove along with a >18 degrees bend in the helix axis toward the major groove. This allosteric perturbation of the DNA helix provides a molecular basis for disruption of transcription factor-DNA interfaces by small molecules, a minimum step in chemical control of gene networks.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
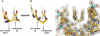
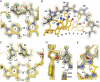
Similar articles
-
Structural basis for cyclic Py-Im polyamide allosteric inhibition of nuclear receptor binding.J Am Chem Soc. 2010 Oct 20;132(41):14521-9. doi: 10.1021/ja105068b. J Am Chem Soc. 2010. PMID: 20812704 Free PMC article.
-
Allosteric analysis of glucocorticoid receptor-DNA interface induced by cyclic Py-Im polyamide: a molecular dynamics simulation study.PLoS One. 2012;7(4):e35159. doi: 10.1371/journal.pone.0035159. Epub 2012 Apr 19. PLoS One. 2012. PMID: 22532842 Free PMC article.
-
RNA polymerase II senses obstruction in the DNA minor groove via a conserved sensor motif.Proc Natl Acad Sci U S A. 2016 Nov 1;113(44):12426-12431. doi: 10.1073/pnas.1612745113. Epub 2016 Oct 17. Proc Natl Acad Sci U S A. 2016. PMID: 27791148 Free PMC article.
-
Sequence-specific DNA binding Pyrrole-imidazole polyamides and their applications.Bioorg Med Chem. 2018 May 1;26(8):1393-1411. doi: 10.1016/j.bmc.2018.01.026. Epub 2018 Feb 1. Bioorg Med Chem. 2018. PMID: 29439914 Review.
-
Recognition of the DNA minor groove by pyrrole-imidazole polyamides.Curr Opin Struct Biol. 2003 Jun;13(3):284-99. doi: 10.1016/s0959-440x(03)00081-2. Curr Opin Struct Biol. 2003. PMID: 12831879 Review.
Cited by
-
B-DNA structure is intrinsically polymorphic: even at the level of base pair positions.Nucleic Acids Res. 2012 Apr;40(8):3714-22. doi: 10.1093/nar/gkr1168. Epub 2011 Dec 17. Nucleic Acids Res. 2012. PMID: 22180536 Free PMC article.
-
Structure-dependent inhibition of the ETS-family transcription factor PU.1 by novel heterocyclic diamidines.Nucleic Acids Res. 2014 Jan;42(2):1379-90. doi: 10.1093/nar/gkt955. Epub 2013 Oct 23. Nucleic Acids Res. 2014. PMID: 24157839 Free PMC article.
-
Enhancing the cellular uptake of Py-Im polyamides through next-generation aryl turns.Nucleic Acids Res. 2012 Mar;40(5):2345-56. doi: 10.1093/nar/gkr970. Epub 2011 Nov 12. Nucleic Acids Res. 2012. PMID: 22080545 Free PMC article.
-
Allosterism and signal transfer in DNA.Nucleic Acids Res. 2018 Sep 6;46(15):7554-7565. doi: 10.1093/nar/gky549. Nucleic Acids Res. 2018. PMID: 29905860 Free PMC article.
-
Tumor Repression of VCaP Xenografts by a Pyrrole-Imidazole Polyamide.PLoS One. 2015 Nov 16;10(11):e0143161. doi: 10.1371/journal.pone.0143161. eCollection 2015. PLoS One. 2015. PMID: 26571387 Free PMC article.
References
-
- Dervan PB. Molecular recognition of DNA by small molecules. Bioorg Med Chem. 2001;9:2215–2235. - PubMed
-
- Dervan PB, Edelson BS. Recognition of the DNA minor groove by pyrrole- imidazole polyamides. Curr Opin Struct Biol. 2003;13:284–299. - PubMed
-
- Trauger JW, Baird EE, Dervan PB. Recognition of DNA by designed ligands at subnanomolar concentrations. Nature. 1996;382:559–561. - PubMed
-
- White S, Szewczyk JW, Turner JM, Baird EE, Dervan PB. Recognition of the four Watson-Crick base pairs in the DNA minor groove by synthetic ligands. Nature. 1998;391:468–470. - PubMed
-
- Kielkopf CL, Baird EE, Dervan PB, Rees DC. Structural basis for G. C recognition in the DNA minor groove. Nat Struct Biol. 1998;5:104–109. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

