Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells
- PMID: 19665978
- PMCID: PMC2731699
- DOI: 10.1016/j.cell.2009.07.011
Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells
Abstract
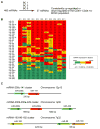
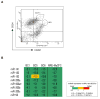
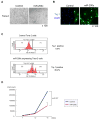
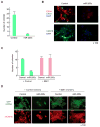
Human breast tumors contain a breast cancer stem cell (BCSC) population with properties reminiscent of normal stem cells. We found 37 microRNAs that were differentially expressed between human BCSCs and nontumorigenic cancer cells. Three clusters, miR-200c-141, miR-200b-200a-429, and miR-183-96-182 were downregulated in human BCSCs, normal human and murine mammary stem/progenitor cells, and embryonal carcinoma cells. Expression of BMI1, a known regulator of stem cell self-renewal, was modulated by miR-200c. miR-200c inhibited the clonal expansion of breast cancer cells and suppressed the growth of embryonal carcinoma cells in vitro. Most importantly, miR-200c strongly suppressed the ability of normal mammary stem cells to form mammary ducts and tumor formation driven by human BCSCs in vivo. The coordinated downregulation of three microRNA clusters and the similar functional regulation of clonal expansion by miR-200c provide a molecular link that connects BCSCs with normal stem cells.
Figures


Comment in
-
MicroRNAs and parallel stem cell lives.Cell. 2009 Aug 7;138(3):423-4. doi: 10.1016/j.cell.2009.07.025. Cell. 2009. PMID: 19665962
Similar articles
-
Essential role of miR-200c in regulating self-renewal of breast cancer stem cells and their counterparts of mammary epithelium.BMC Cancer. 2015 Sep 23;15:645. doi: 10.1186/s12885-015-1655-5. BMC Cancer. 2015. PMID: 26400441 Free PMC article.
-
MicroRNAs and parallel stem cell lives.Cell. 2009 Aug 7;138(3):423-4. doi: 10.1016/j.cell.2009.07.025. Cell. 2009. PMID: 19665962
-
miR-200c/141 Regulates Breast Cancer Stem Cell Heterogeneity via Targeting HIPK1/β-Catenin Axis.Theranostics. 2018 Nov 10;8(21):5801-5813. doi: 10.7150/thno.29380. eCollection 2018. Theranostics. 2018. PMID: 30613263 Free PMC article.
-
Epithelial-mesenchymal transition and cancer stemness: the Twist1-Bmi1 connection.Biosci Rep. 2011 Dec;31(6):449-55. doi: 10.1042/BSR20100114. Biosci Rep. 2011. PMID: 21919891 Review.
-
The ups and downs of miR-205: identifying the roles of miR-205 in mammary gland development and breast cancer.RNA Biol. 2010 May-Jun;7(3):300-4. doi: 10.4161/rna.7.3.11837. Epub 2010 May 21. RNA Biol. 2010. PMID: 20436283 Free PMC article. Review.
Cited by
-
FGFR2 promotes breast tumorigenicity through maintenance of breast tumor-initiating cells.PLoS One. 2013;8(1):e51671. doi: 10.1371/journal.pone.0051671. Epub 2013 Jan 2. PLoS One. 2013. PMID: 23300950 Free PMC article.
-
Downregulation of miR-140 promotes cancer stem cell formation in basal-like early stage breast cancer.Oncogene. 2014 May 15;33(20):2589-600. doi: 10.1038/onc.2013.226. Epub 2013 Jun 10. Oncogene. 2014. PMID: 23752191 Free PMC article.
-
Adipsin-dependent adipocyte maturation induces cancer cell invasion in breast cancer.Sci Rep. 2024 Aug 9;14(1):18494. doi: 10.1038/s41598-024-69476-3. Sci Rep. 2024. PMID: 39122742 Free PMC article.
-
Roles and epigenetic regulation of epithelial-mesenchymal transition and its transcription factors in cancer initiation and progression.Cell Mol Life Sci. 2016 Dec;73(24):4643-4660. doi: 10.1007/s00018-016-2313-z. Epub 2016 Jul 26. Cell Mol Life Sci. 2016. PMID: 27460000 Free PMC article. Review.
-
A novel miR-200b-3p/p38IP pair regulates monocyte/macrophage differentiation.Cell Discov. 2016 Jan 26;2:15043. doi: 10.1038/celldisc.2015.43. eCollection 2016. Cell Discov. 2016. PMID: 27462440 Free PMC article.
References
-
- Akala OO, Park IK, Qian D, Pihalja M, Becker MW, Clarke MF. Long-term haematopoietic reconstitution by Trp53-/-p16Ink4a-/-p19Arf-/- multipotent progenitors. Nature. 2008;453:228–232. - PubMed
-
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. - PubMed
-
- Bruce WR, Gaag H. A quantitative assay for the number of murine lymphoma cells capable of proliferation in vivo. Nature. 1963;199:79–80. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

