Role for G-quadruplex RNA binding by Epstein-Barr virus nuclear antigen 1 in DNA replication and metaphase chromosome attachment
- PMID: 19656898
- PMCID: PMC2753104
- DOI: 10.1128/JVI.00747-09
Role for G-quadruplex RNA binding by Epstein-Barr virus nuclear antigen 1 in DNA replication and metaphase chromosome attachment
Abstract
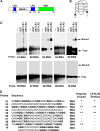
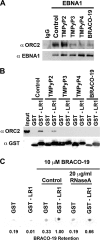
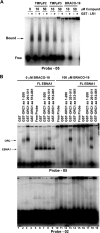
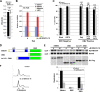
Latent infection by Epstein-Barr virus (EBV) requires both replication and maintenance of the viral genome. EBV nuclear antigen 1 (EBNA1) is a virus-encoded protein that is critical for the replication and maintenance of the genome during latency in proliferating cells. We have previously demonstrated that EBNA1 recruits the cellular origin recognition complex (ORC) through an RNA-dependent interaction with EBNA1 linking region 1 (LR1) and LR2. We now show that LR1 and LR2 bind to G-rich RNA that is predicted to form G-quadruplex structures. Several chemically distinct G-quadruplex-interacting drugs disrupted the interaction between EBNA1 and ORC. The G-quadruplex-interacting compound BRACO-19 inhibited EBNA1-dependent stimulation of viral DNA replication and preferentially blocked proliferation of EBV-positive cells relative to EBV-negative cell lines. BRACO-19 treatment also disrupted the ability of EBNA1 to tether to metaphase chromosomes, suggesting that maintenance function is also mediated through G-quadruplex recognition. These findings suggest that the EBNA1 replication and maintenance function uses a common G-quadruplex binding capacity of LR1 and LR2, which may be targetable by small-molecule inhibitors.
Figures
Similar articles
-
Structural and Functional Basis for an EBNA1 Hexameric Ring in Epstein-Barr Virus Episome Maintenance.J Virol. 2017 Sep 12;91(19):e01046-17. doi: 10.1128/JVI.01046-17. Print 2017 Oct 1. J Virol. 2017. PMID: 28701406 Free PMC article.
-
Structural Basis for Cooperative Binding of EBNA1 to the Epstein-Barr Virus Dyad Symmetry Minimal Origin of Replication.J Virol. 2019 Sep 30;93(20):e00487-19. doi: 10.1128/JVI.00487-19. Print 2019 Oct 15. J Virol. 2019. PMID: 31142669 Free PMC article.
-
The amino terminus of Epstein-Barr Virus (EBV) nuclear antigen 1 contains AT hooks that facilitate the replication and partitioning of latent EBV genomes by tethering them to cellular chromosomes.J Virol. 2004 Nov;78(21):11487-505. doi: 10.1128/JVI.78.21.11487-11505.2004. J Virol. 2004. PMID: 15479791 Free PMC article.
-
The 2.2 A structure of a permanganate-sensitive DNA site bound by the Epstein-Barr virus origin binding protein, EBNA1.J Mol Biol. 1998 Dec 18;284(5):1273-8. doi: 10.1006/jmbi.1998.2247. J Mol Biol. 1998. PMID: 9878348 Review.
-
Replication licensing of the EBV oriP minichromosome.Curr Top Microbiol Immunol. 2001;258:13-33. doi: 10.1007/978-3-642-56515-1_2. Curr Top Microbiol Immunol. 2001. PMID: 11443858 Review.
Cited by
-
Major Achievements in the Design of Quadruplex-Interactive Small Molecules.Pharmaceuticals (Basel). 2022 Feb 28;15(3):300. doi: 10.3390/ph15030300. Pharmaceuticals (Basel). 2022. PMID: 35337098 Free PMC article. Review.
-
Genome-wide analysis of host-chromosome binding sites for Epstein-Barr Virus Nuclear Antigen 1 (EBNA1).Virol J. 2010 Oct 7;7:262. doi: 10.1186/1743-422X-7-262. Virol J. 2010. PMID: 20929547 Free PMC article.
-
Identification of MEF2B, EBF1, and IL6R as Direct Gene Targets of Epstein-Barr Virus (EBV) Nuclear Antigen 1 Critical for EBV-Infected B-Lymphocyte Survival.J Virol. 2015 Oct 14;90(1):345-55. doi: 10.1128/JVI.02318-15. Print 2016 Jan 1. J Virol. 2015. PMID: 26468528 Free PMC article.
-
Synthesis, Binding and Antiviral Properties of Potent Core-Extended Naphthalene Diimides Targeting the HIV-1 Long Terminal Repeat Promoter G-Quadruplexes.J Med Chem. 2015 Dec 24;58(24):9639-52. doi: 10.1021/acs.jmedchem.5b01283. Epub 2015 Dec 8. J Med Chem. 2015. PMID: 26599611 Free PMC article.
-
Kaposi's Sarcoma-Associated Herpesvirus Genome Replication, Partitioning, and Maintenance in Latency.Front Microbiol. 2012 Jan 24;3:7. doi: 10.3389/fmicb.2012.00007. eCollection 2012. Front Microbiol. 2012. PMID: 22291692 Free PMC article.
References
-
- Bochkarev, A., J. A. Barwell, R. A. Pfuetzner, E. Bochkareva, L. Frappier, and A. M. Edwards. 1996. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein, EBNA1, bound to DNA. Cell 84:791-800. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

