The organization of the transcriptional network in specific neuronal classes
- PMID: 19638972
- PMCID: PMC2724976
- DOI: 10.1038/msb.2009.46
The organization of the transcriptional network in specific neuronal classes
Abstract
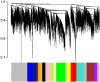
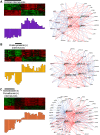
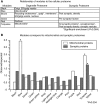
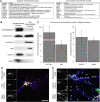
Genome-wide expression profiling has aided the understanding of the molecular basis of neuronal diversity, but achieving broad functional insight remains a considerable challenge. Here, we perform the first systems-level analysis of microarray data from single neuronal populations using weighted gene co-expression network analysis to examine how neuronal transcriptome organization relates to neuronal function and diversity. We systematically validate network predictions using published proteomic and genomic data. Several network modules of co-expressed genes correspond to interneuron development programs, in which the hub genes are known to be critical for interneuron specification. Other co-expression modules relate to fundamental cellular functions, such as energy production, firing rate, trafficking, and synapses, suggesting that fundamental aspects of neuronal diversity are produced by quantitative variation in basic metabolic processes. We identify two transcriptionally distinct mitochondrial modules and demonstrate that one corresponds to mitochondria enriched in neuronal processes and synapses, whereas the other represents a population restricted to the soma. Finally, we show that galectin-1 is a new interneuron marker, and we validate network predictions in vivo using Rgs4 and Dlx1/2 knockout mice. These analyses provide a basis for understanding how specific aspects of neuronal phenotypic diversity are organized at the transcriptional level.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Similar articles
-
Co-expression network analysis identified hub genes critical to triglyceride and free fatty acid metabolism as key regulators of age-related vascular dysfunction in mice.Aging (Albany NY). 2019 Sep 12;11(18):7620-7638. doi: 10.18632/aging.102275. Epub 2019 Sep 12. Aging (Albany NY). 2019. PMID: 31514170 Free PMC article.
-
Arabidopsis gene co-expression network and its functional modules.BMC Bioinformatics. 2009 Oct 21;10:346. doi: 10.1186/1471-2105-10-346. BMC Bioinformatics. 2009. PMID: 19845953 Free PMC article.
-
Identifying gene modules of thyroid cancer associated with pathological stage by weighted gene co-expression network analysis.Gene. 2019 Jul 1;704:142-148. doi: 10.1016/j.gene.2019.04.017. Epub 2019 Apr 6. Gene. 2019. PMID: 30965127
-
Identification of critical genes associated with the development of asthma by co-expression modules construction.Mol Immunol. 2020 Jul;123:18-25. doi: 10.1016/j.molimm.2020.01.015. Epub 2020 May 6. Mol Immunol. 2020. PMID: 32388106
-
Co-expression networks for plant biology: why and how.Acta Biochim Biophys Sin (Shanghai). 2019 Sep 6;51(10):981-988. doi: 10.1093/abbs/gmz080. Acta Biochim Biophys Sin (Shanghai). 2019. PMID: 31436787 Review.
Cited by
-
Cell type-specific genes show striking and distinct patterns of spatial expression in the mouse brain.Proc Natl Acad Sci U S A. 2013 Feb 19;110(8):3095-100. doi: 10.1073/pnas.1222897110. Epub 2013 Feb 5. Proc Natl Acad Sci U S A. 2013. PMID: 23386717 Free PMC article.
-
Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits.Cell. 2011 Sep 30;147(1):235-46. doi: 10.1016/j.cell.2011.08.040. Cell. 2011. PMID: 21962519 Free PMC article.
-
Accelerated brain aging towards transcriptional inversion in a zebrafish model of the K115fs mutation of human PSEN2.PLoS One. 2020 Jan 24;15(1):e0227258. doi: 10.1371/journal.pone.0227258. eCollection 2020. PLoS One. 2020. PMID: 31978074 Free PMC article.
-
Evolution, immunity and the emergence of brain superautoantigens.F1000Res. 2017 Feb 21;6:171. doi: 10.12688/f1000research.10950.1. eCollection 2017. F1000Res. 2017. PMID: 28529699 Free PMC article.
-
Amylin Treatment Reduces Neuroinflammation and Ameliorates Abnormal Patterns of Gene Expression in the Cerebral Cortex of an Alzheimer's Disease Mouse Model.J Alzheimers Dis. 2017;56(1):47-61. doi: 10.3233/JAD-160677. J Alzheimers Dis. 2017. PMID: 27911303 Free PMC article.
References
-
- Albert R, Jeong H, Barabasi A (2000) Error and attack tolerance of complex networks. Nature 406: 378–382 - PubMed
-
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JL (1997a) Interneuron migration from basal forebrain to neocortex: dependence on dlx genes. Science 278: 474–476 - PubMed
-
- Anderson SA, Qiu M, Bulfone A, Eisenstat DD, Meneses J, Pedersen R, Rubenstein JL (1997b) Mutations of the homeobox genes dlx-1 and dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron 19: 27–37 - PubMed
-
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD (2005) Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron 45: 207–221 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

