Bridging IRES elements in mRNAs to the eukaryotic translation apparatus
- PMID: 19631772
- PMCID: PMC2783899
- DOI: 10.1016/j.bbagrm.2009.07.004
Bridging IRES elements in mRNAs to the eukaryotic translation apparatus
Abstract
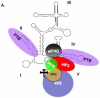
IRES elements are highly structured RNA sequences that function to recruit ribosomes for the initiation of translation. In contrast to the canonical cap-binding, ribosome-scanning model, the mechanism of IRES-mediated translation initiation is not well understood. IRES elements, first discovered in viral RNA genomes, were subsequently found in a subset of cellular RNAs as well. Interestingly, these cellular IRES-containing mRNAs appear to play important roles during conditions of cellular stress, development, and disease (e.g., cancer). It has been shown for viral IRESes that some require specific IRES trans-acting factors (ITAFs), while others require few if any additional proteins and can bind ribosomes directly. Current studies are aimed at elucidating the mechanism of IRES-mediated translation initiation and features that may be common or differ greatly among cellular and viral IRESes. This review will explore IRES elements as important RNA structures that function in both cellular and viral RNA translation and the significance of these structures in providing an alternative mechanism of eukaryotic translation initiation.
Figures

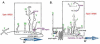


Similar articles
-
Advances and Breakthroughs in IRES-Directed Translation and Replication of Picornaviruses.mBio. 2023 Apr 25;14(2):e0035823. doi: 10.1128/mbio.00358-23. Epub 2023 Mar 20. mBio. 2023. PMID: 36939331 Free PMC article. Review.
-
Eukaryotic initiation factor 4G-poly(A) binding protein interaction is required for poly(A) tail-mediated stimulation of picornavirus internal ribosome entry segment-driven translation but not for X-mediated stimulation of hepatitis C virus translation.Mol Cell Biol. 2001 Jul;21(13):4097-109. doi: 10.1128/MCB.21.13.4097-4109.2001. Mol Cell Biol. 2001. PMID: 11390639 Free PMC article.
-
Picornavirus IRES elements: RNA structure and host protein interactions.Virus Res. 2015 Aug 3;206:62-73. doi: 10.1016/j.virusres.2015.01.012. Epub 2015 Jan 21. Virus Res. 2015. PMID: 25617758 Review.
-
Internal translation initiation of picornaviruses and hepatitis C virus.Biochim Biophys Acta. 2009 Sep-Oct;1789(9-10):529-41. doi: 10.1016/j.bbagrm.2009.05.002. Epub 2009 May 9. Biochim Biophys Acta. 2009. PMID: 19439208 Review.
-
Insights into the biology of IRES elements through riboproteomic approaches.J Biomed Biotechnol. 2010;2010:458927. doi: 10.1155/2010/458927. Epub 2010 Feb 2. J Biomed Biotechnol. 2010. PMID: 20150968 Free PMC article. Review.
Cited by
-
Functional and structural analysis of the internal ribosome entry site present in the mRNA of natural variants of the HIV-1.PLoS One. 2012;7(4):e35031. doi: 10.1371/journal.pone.0035031. Epub 2012 Apr 4. PLoS One. 2012. PMID: 22496887 Free PMC article.
-
Kinetics of initiating polypeptide elongation in an IRES-dependent system.Elife. 2016 Jun 2;5:e13429. doi: 10.7554/eLife.13429. Elife. 2016. PMID: 27253065 Free PMC article.
-
Additive Promotion of Viral Internal Ribosome Entry Site-Mediated Translation by Far Upstream Element-Binding Protein 1 and an Enterovirus 71-Induced Cleavage Product.PLoS Pathog. 2016 Oct 25;12(10):e1005959. doi: 10.1371/journal.ppat.1005959. eCollection 2016 Oct. PLoS Pathog. 2016. PMID: 27780225 Free PMC article.
-
SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage.PLoS Pathog. 2011 Dec;7(12):e1002433. doi: 10.1371/journal.ppat.1002433. Epub 2011 Dec 8. PLoS Pathog. 2011. PMID: 22174690 Free PMC article.
-
So close, no matter how far: multiple paths connecting transcription to mRNA translation in eukaryotes.EMBO Rep. 2020 Sep 3;21(9):e50799. doi: 10.15252/embr.202050799. Epub 2020 Aug 16. EMBO Rep. 2020. PMID: 32803873 Free PMC article. Review.
References
-
- Jackson RJ. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem. Soc. Trans. 2005;33:1231. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

