Respiratory syncytial virus nonstructural proteins decrease levels of multiple members of the cellular interferon pathways
- PMID: 19625398
- PMCID: PMC2748017
- DOI: 10.1128/JVI.00715-09
Respiratory syncytial virus nonstructural proteins decrease levels of multiple members of the cellular interferon pathways
Abstract
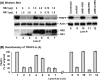
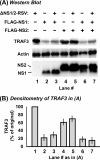
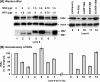
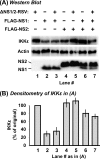
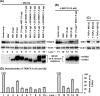
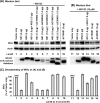
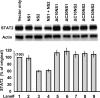
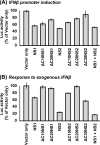
Viruses of the Paramyxoviridae family, such as the respiratory syncytial virus (RSV), suppress cellular innate immunity represented by type I interferon (IFN) for optimal growth in their hosts. The two unique nonstructural (NS) proteins, NS1 and NS2, of RSV suppress IFN synthesis, as well as IFN function, but their exact targets are still uncharacterized. Here, we investigate if either or both of the NS proteins affect the steady-state levels of key members of the IFN pathway. We found that both NS1 and NS2 decreased the levels of TRAF3, a strategic integrator of multiple IFN-inducing signals, although NS1 was more efficient. Only NS1 reduced IKKepsilon, a key protein kinase that specifically phosphorylates and activates IFN regulatory factor 3. Loss of the TRAF3 and IKKepsilon proteins appeared to involve a nonproteasomal mechanism. Interestingly, NS2 modestly increased IKKepsilon levels. In the IFN response pathway, NS2 decreased the levels of STAT2, the essential transcription factor for IFN-inducible antiviral genes. Preliminary mapping revealed that the C-terminal 10 residues of NS1 were essential for reducing IKKepsilon levels and the C-terminal 10 residues of NS2 were essential for increasing and reducing IKKepsilon and STAT2, respectively. In contrast, deletion of up to 20 residues of the C termini of NS1 and NS2 did not diminish their TRAF3-reducing activity. Coimmunoprecipitation studies revealed that NS1 and NS2 form a heterodimer. Clearly, the NS proteins of RSV, working individually and together, regulate key signaling molecules of both the IFN activation and response pathways.
Figures

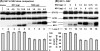
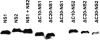


Similar articles
-
Multiple functional domains and complexes of the two nonstructural proteins of human respiratory syncytial virus contribute to interferon suppression and cellular location.J Virol. 2011 Oct;85(19):10090-100. doi: 10.1128/JVI.00413-11. Epub 2011 Jul 27. J Virol. 2011. PMID: 21795342 Free PMC article.
-
Effects of nonstructural proteins NS1 and NS2 of human respiratory syncytial virus on interferon regulatory factor 3, NF-kappaB, and proinflammatory cytokines.J Virol. 2005 May;79(9):5353-62. doi: 10.1128/JVI.79.9.5353-5362.2005. J Virol. 2005. PMID: 15827150 Free PMC article.
-
Identification of Respiratory Syncytial Virus Nonstructural Protein 2 Residues Essential for Exploitation of the Host Ubiquitin System and Inhibition of Innate Immune Responses.J Virol. 2016 Jun 24;90(14):6453-6463. doi: 10.1128/JVI.00423-16. Print 2016 Jul 15. J Virol. 2016. PMID: 27147743 Free PMC article.
-
Respiratory syncytial virus nonstructural proteins 1 and 2: Exceptional disrupters of innate immune responses.PLoS Pathog. 2019 Oct 17;15(10):e1007984. doi: 10.1371/journal.ppat.1007984. eCollection 2019 Oct. PLoS Pathog. 2019. PMID: 31622448 Free PMC article. Review.
-
Respiratory syncytial virus mechanisms to interfere with type 1 interferons.Curr Top Microbiol Immunol. 2013;372:173-91. doi: 10.1007/978-3-642-38919-1_9. Curr Top Microbiol Immunol. 2013. PMID: 24362690 Review.
Cited by
-
Identification of Interferon-Stimulated Gene Proteins That Inhibit Human Parainfluenza Virus Type 3.J Virol. 2016 Nov 28;90(24):11145-11156. doi: 10.1128/JVI.01551-16. Print 2016 Dec 15. J Virol. 2016. PMID: 27707917 Free PMC article.
-
Induction of interferon-stimulated genes by IRF3 promotes replication of Toxoplasma gondii.PLoS Pathog. 2015 Mar 26;11(3):e1004779. doi: 10.1371/journal.ppat.1004779. eCollection 2015 Mar. PLoS Pathog. 2015. Retraction in: PLoS Pathog. 2024 May 31;20(5):e1012264. doi: 10.1371/journal.ppat.1012264 PMID: 25811886 Free PMC article. Retracted.
-
Mechanisms of Viral Degradation of Cellular Signal Transducer and Activator of Transcription 2.Int J Mol Sci. 2022 Jan 1;23(1):489. doi: 10.3390/ijms23010489. Int J Mol Sci. 2022. PMID: 35008916 Free PMC article. Review.
-
A comprehensive proteomic view of responses of A549 type II alveolar epithelial cells to human respiratory syncytial virus infection.Mol Cell Proteomics. 2014 Dec;13(12):3250-69. doi: 10.1074/mcp.M114.041129. Epub 2014 Aug 8. Mol Cell Proteomics. 2014. PMID: 25106423 Free PMC article.
-
Respiratory Syncytial Virus Persistence in Murine Macrophages Impairs IFN-β Response but Not Synthesis.Viruses. 2015 Oct 16;7(10):5361-74. doi: 10.3390/v7102879. Viruses. 2015. PMID: 26501312 Free PMC article.
References
-
- Bitko, V., O. Shulyayeva, B. Mazumder, A. Musiyenko, M. Ramaswamy, D. C. Look, and S. Barik. 2007. Nonstructural proteins of respiratory syncytial virus suppress premature apoptosis by an NF-κB-dependent, interferon-independent mechanism and facilitate virus growth. J. Virol. 81:1786-1795. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

