Evolution and survival on eutherian sex chromosomes
- PMID: 19609352
- PMCID: PMC2704370
- DOI: 10.1371/journal.pgen.1000568
Evolution and survival on eutherian sex chromosomes
Abstract
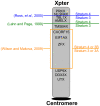
Since the two eutherian sex chromosomes diverged from an ancestral autosomal pair, the X has remained relatively gene-rich, while the Y has lost most of its genes through the accumulation of deleterious mutations in nonrecombining regions. Presently, it is unclear what is distinctive about genes that remain on the Y chromosome, when the sex chromosomes acquired their unique evolutionary rates, and whether X-Y gene divergence paralleled that of paralogs located on autosomes. To tackle these questions, here we juxtaposed the evolution of X and Y homologous genes (gametologs) in eutherian mammals with their autosomal orthologs in marsupial and monotreme mammals. We discovered that genes on the X and Y acquired distinct evolutionary rates immediately following the suppression of recombination between the two sex chromosomes. The Y-linked genes evolved at higher rates, while the X-linked genes maintained the lower evolutionary rates of the ancestral autosomal genes. These distinct rates have been maintained throughout the evolution of X and Y. Specifically, in humans, most X gametologs and, curiously, also most Y gametologs evolved under stronger purifying selection than similarly aged autosomal paralogs. Finally, after evaluating the current experimental data from the literature, we concluded that unique mRNA/protein expression patterns and functions acquired by Y (versus X) gametologs likely contributed to their retention. Our results also suggest that either the boundary between sex chromosome strata 3 and 4 should be shifted or that stratum 3 should be divided into two strata.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
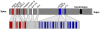
Similar articles
-
No evidence for a second evolutionary stratum during the early evolution of mammalian sex chromosomes.PLoS One. 2012;7(10):e45488. doi: 10.1371/journal.pone.0045488. Epub 2012 Oct 19. PLoS One. 2012. PMID: 23094017 Free PMC article.
-
A gene deriving from the ancestral sex chromosomes was lost from the X and retained on the Y chromosome in eutherian mammals.BMC Biol. 2022 Jun 9;20(1):133. doi: 10.1186/s12915-022-01338-8. BMC Biol. 2022. PMID: 35676717 Free PMC article.
-
Shared DNA sequences between the X and Y chromosomes in the tammar wallaby - evidence for independent additions to eutherian and marsupial sex chromosomes.Chromosoma. 1997 Jul;106(2):94-8. doi: 10.1007/s004120050228. Chromosoma. 1997. PMID: 9215558
-
Reconstructing the evolution of vertebrate sex chromosomes.Cold Spring Harb Symp Quant Biol. 2009;74:345-53. doi: 10.1101/sqb.2009.74.048. Epub 2010 May 27. Cold Spring Harb Symp Quant Biol. 2009. PMID: 20508063 Review.
-
The origin and evolution of the pseudoautosomal regions of human sex chromosomes.Hum Mol Genet. 1998 Dec;7(13):1991-6. doi: 10.1093/hmg/7.13.1991. Hum Mol Genet. 1998. PMID: 9817914 Review.
Cited by
-
Genomic and demographic processes differentially influence genetic variation across the human X chromosome.PLoS One. 2023 Nov 1;18(11):e0287609. doi: 10.1371/journal.pone.0287609. eCollection 2023. PLoS One. 2023. PMID: 37910456 Free PMC article.
-
Chromosome-Aware Phylogenomics of Assassin Bugs (Hemiptera: Reduvioidea) Elucidates Ancient Gene Conflict.Mol Biol Evol. 2023 Aug 3;40(8):msad168. doi: 10.1093/molbev/msad168. Mol Biol Evol. 2023. PMID: 37494292 Free PMC article.
-
Consequence of evolutionary loss of seasonal breeding by humans for prostate cancer chemoprevention.Am J Clin Exp Urol. 2023 Jun 15;11(3):194-205. eCollection 2023. Am J Clin Exp Urol. 2023. PMID: 37441442 Free PMC article. Review.
-
Structural shifts in primate Y.Nat Ecol Evol. 2023 Jul;7(7):971-972. doi: 10.1038/s41559-023-01984-3. Nat Ecol Evol. 2023. PMID: 37268855 No abstract available.
-
Sex differences in early and term placenta are conserved in adult tissues.Biol Sex Differ. 2022 Dec 22;13(1):74. doi: 10.1186/s13293-022-00470-y. Biol Sex Differ. 2022. PMID: 36550527 Free PMC article.
References
-
- Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. - PubMed
-
- Graves JAM. Sex chromosome specialization and degeneration in mammals. Cell. 2006;124:901–914. - PubMed
-
- Wallis MC, Waters PD, Delbridge ML, Kirby PJ, Pask AJ, et al. Sex determination in platypus and echidna: autosomal location of SOX3 confirms the absence of SRY from monotremes. Chromosome Res. 2007;15:949–959. - PubMed
-
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

