Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination
- PMID: 19595722
- PMCID: PMC2746756
- DOI: 10.1016/j.molcel.2009.06.019
Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination
Abstract
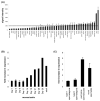
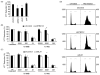
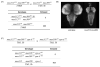
DNA recombination and repair pathways require structure-specific endonucleases to process DNA structures that include forks, flaps, and Holliday junctions. Previously, we determined that the Drosophila MEI-9-ERCC1 endonuclease interacts with the MUS312 protein to produce meiotic crossovers, and that MUS312 has a MEI-9-independent role in interstrand crosslink (ICL) repair. The importance of MUS312 to pathways crucial for maintaining genomic stability in Drosophila prompted us to search for orthologs in other organisms. Based on sequence, expression pattern, conserved protein-protein interactions, and ICL repair function, we determined that the mammalian ortholog of MUS312 is BTBD12. Orthology between these proteins and S. cerevisiae Slx4 helped identify a conserved interaction with a second structure-specific endonuclease, SLX1. Genetic and biochemical evidence described here and in related papers suggest that MUS312 and BTBD12 direct Holliday junction resolution by at least two distinct endonucleases in different recombination and repair contexts.
Figures

Similar articles
-
Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases.Cell. 2009 Jul 10;138(1):78-89. doi: 10.1016/j.cell.2009.06.029. Cell. 2009. PMID: 19596236 Free PMC article.
-
Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair.Mol Cell. 2009 Jul 10;35(1):116-27. doi: 10.1016/j.molcel.2009.06.020. Mol Cell. 2009. PMID: 19595721
-
Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair.Cell. 2009 Jul 10;138(1):63-77. doi: 10.1016/j.cell.2009.06.030. Cell. 2009. PMID: 19596235 Free PMC article.
-
Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes.Chromosoma. 2011 Apr;120(2):109-27. doi: 10.1007/s00412-010-0304-7. Epub 2011 Jan 11. Chromosoma. 2011. PMID: 21369956 Free PMC article. Review.
-
Orchestrating the nucleases involved in DNA interstrand cross-link (ICL) repair.Cell Cycle. 2011 Dec 1;10(23):3999-4008. doi: 10.4161/cc.10.23.18385. Epub 2011 Dec 1. Cell Cycle. 2011. PMID: 22101340 Free PMC article. Review.
Cited by
-
DNA repair endonuclease ERCC1-XPF as a novel therapeutic target to overcome chemoresistance in cancer therapy.Nucleic Acids Res. 2012 Nov 1;40(20):9990-10004. doi: 10.1093/nar/gks818. Epub 2012 Aug 31. Nucleic Acids Res. 2012. PMID: 22941649 Free PMC article. Review.
-
The DNA damage response: making it safe to play with knives.Mol Cell. 2010 Oct 22;40(2):179-204. doi: 10.1016/j.molcel.2010.09.019. Mol Cell. 2010. PMID: 20965415 Free PMC article. Review.
-
Meiotic checkpoints and the interchromosomal effect on crossing over in Drosophila females.Fly (Austin). 2011 Apr-Jun;5(2):134-40. doi: 10.4161/fly.5.2.14767. Epub 2011 Apr 1. Fly (Austin). 2011. PMID: 21339705 Free PMC article.
-
Three structure-selective endonucleases are essential in the absence of BLM helicase in Drosophila.PLoS Genet. 2011 Oct;7(10):e1002315. doi: 10.1371/journal.pgen.1002315. Epub 2011 Oct 13. PLoS Genet. 2011. PMID: 22022278 Free PMC article.
-
The SMX DNA Repair Tri-nuclease.Mol Cell. 2017 Mar 2;65(5):848-860.e11. doi: 10.1016/j.molcel.2017.01.031. Mol Cell. 2017. PMID: 28257701 Free PMC article.
References
-
- Aravind L, Koonin EV. SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci. 2000;25:112–114. - PubMed
-
- Bardwell AJ, Bardwell L, Tomkinson AE, Friedberg EC. Specific cleavage of model recombination and repair intermediates by the yeast Rad1-Rad10 DNA endonuclease. Science. 1994;265:2082–2085. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

