The zinc finger DNA-binding domain of K-RBP plays an important role in regulating Kaposi's sarcoma-associated herpesvirus RTA-mediated gene expression
- PMID: 19592062
- PMCID: PMC2767234
- DOI: 10.1016/j.virol.2009.06.014
The zinc finger DNA-binding domain of K-RBP plays an important role in regulating Kaposi's sarcoma-associated herpesvirus RTA-mediated gene expression
Abstract
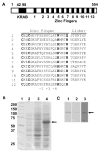
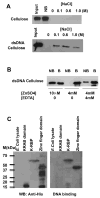
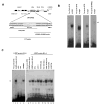
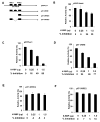
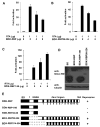
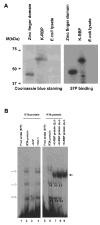
K-RBP is a KRAB-containing zinc finger protein with multiple zinc finger motifs and represses Kaposi's sarcoma-associated herpesvirus (KSHV) transactivator RTA-mediated transactivation of several viral lytic gene promoters, including the ORF57 promoter. Whether K-RBP binds DNA through its zinc fingers and the role of zinc finger domain in repressing gene expression are unclear. Here we report that K-RBP binds DNA through its zinc finger domain and the target DNA sequences contain high GC content. Furthermore, K-RBP binds to KSHV ORF57 promoter, which contains a GC-rich motif. K-RBP suppresses the basal ORF57 promoter activity as well as RTA-mediated activation. The zinc finger domain of K-RBP is sufficient for the suppression of ORF57 promoter activation mediated by the viral transactivator RTA. Finally, we show that K-RBP inhibits RTA binding to ORF57 promoter. These findings suggest that the DNA-binding activity of K-RBP plays an important role in repressing viral promoter activity.
Figures

Similar articles
-
The transcriptional repressor K-RBP modulates RTA-mediated transactivation and lytic replication of Kaposi's sarcoma-associated herpesvirus.J Virol. 2007 Jun;81(12):6294-306. doi: 10.1128/JVI.02648-06. Epub 2007 Apr 4. J Virol. 2007. PMID: 17409159 Free PMC article.
-
Direct interactions of Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 ORF50/Rta protein with the cellular protein octamer-1 and DNA are critical for specifying transactivation of a delayed-early promoter and stimulating viral reactivation.J Virol. 2007 Aug;81(16):8451-67. doi: 10.1128/JVI.00265-07. Epub 2007 May 30. J Virol. 2007. PMID: 17537858 Free PMC article.
-
NF-kappaB serves as a cellular sensor of Kaposi's sarcoma-associated herpesvirus latency and negatively regulates K-Rta by antagonizing the RBP-Jkappa coactivator.J Virol. 2009 May;83(9):4435-46. doi: 10.1128/JVI.01999-08. Epub 2009 Feb 25. J Virol. 2009. PMID: 19244329 Free PMC article.
-
The role of Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 regulator of transcription activation (RTA) in control of gene expression.Oncogene. 2003 Aug 11;22(33):5150-63. doi: 10.1038/sj.onc.1206555. Oncogene. 2003. PMID: 12910252 Review.
-
RBM22, a Key Player of Pre-mRNA Splicing and Gene Expression Regulation, Is Altered in Cancer.Cancers (Basel). 2022 Jan 27;14(3):643. doi: 10.3390/cancers14030643. Cancers (Basel). 2022. PMID: 35158909 Free PMC article. Review.
Cited by
-
NCOA2 promotes lytic reactivation of Kaposi's sarcoma-associated herpesvirus by enhancing the expression of the master switch protein RTA.PLoS Pathog. 2019 Nov 21;15(11):e1008160. doi: 10.1371/journal.ppat.1008160. eCollection 2019 Nov. PLoS Pathog. 2019. PMID: 31751430 Free PMC article.
-
Regulation of KSHV Latency and Lytic Reactivation.Viruses. 2020 Sep 17;12(9):1034. doi: 10.3390/v12091034. Viruses. 2020. PMID: 32957532 Free PMC article. Review.
-
Number of and distance between response elements in Kaposi's sarcoma-associated herpesvirus ORF57 promoter influence its activation by replication and transcription activator and its repression by interferon regulatory factor 7.Arch Virol. 2010 Mar;155(3):361-6. doi: 10.1007/s00705-009-0576-5. Epub 2009 Dec 29. Arch Virol. 2010. PMID: 20039088 Free PMC article.
-
Mechanisms of Kaposi's Sarcoma-Associated Herpesvirus Latency and Reactivation.Adv Virol. 2011;2011:193860. doi: 10.1155/2011/193860. Adv Virol. 2011. PMID: 21625290 Free PMC article.
-
Transgene regulation using the tetracycline-inducible TetR-KRAB system after AAV-mediated gene transfer in rodents and nonhuman primates.PLoS One. 2014 Sep 23;9(9):e102538. doi: 10.1371/journal.pone.0102538. eCollection 2014. PLoS One. 2014. PMID: 25248159 Free PMC article.
References
-
- Blackwell TK, Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990;250 (4984):1104–1110. - PubMed
-
- Brayer KJ, Segal DJ. Keep your fingers off my DNA: protein-protein interactions mediated by C2H2 zinc finger domains. Cell Biochem Biophys. 2008;50(3):111–131. - PubMed
-
- Bulliard Y, Wiznerowicz M, Barde I, Trono D. KRAB can repress lentivirus proviral transcription independently of integration site. J Biol Chem. 2006;281(47):35742–35746. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P20 RR015635-01/RR/NCRR NIH HHS/United States
- R01 CA075903/CA/NCI NIH HHS/United States
- R01 CA075903-10/CA/NCI NIH HHS/United States
- R01 CA075903-07/CA/NCI NIH HHS/United States
- K12 CA076903/CA/NCI NIH HHS/United States
- P20 RR015635-09/RR/NCRR NIH HHS/United States
- R01 CA075903-06A1/CA/NCI NIH HHS/United States
- R01 CA075903-10S1/CA/NCI NIH HHS/United States
- R01 CA075903-08/CA/NCI NIH HHS/United States
- P20 RR015635-076596/RR/NCRR NIH HHS/United States
- R01 CA075903-09/CA/NCI NIH HHS/United States
- P20 RR015635-06/RR/NCRR NIH HHS/United States
- P20 RR015635-10/RR/NCRR NIH HHS/United States
- CA76903/CA/NCI NIH HHS/United States
- P20 RR015635-096070/RR/NCRR NIH HHS/United States
- P20 RR015635-07/RR/NCRR NIH HHS/United States
- RR15635/RR/NCRR NIH HHS/United States
- P20 RR015635-086852/RR/NCRR NIH HHS/United States
- P20 RR015635-08/RR/NCRR NIH HHS/United States
- P20 RR015635-059004/RR/NCRR NIH HHS/United States
- P20 RR015635/RR/NCRR NIH HHS/United States
- P20 RR015635-058495/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

