A 49-kilodalton isoform of the adenovirus type 5 early region 1B 55-kilodalton protein is sufficient to support virus replication
- PMID: 19587039
- PMCID: PMC2738261
- DOI: 10.1128/JVI.00728-09
A 49-kilodalton isoform of the adenovirus type 5 early region 1B 55-kilodalton protein is sufficient to support virus replication
Abstract
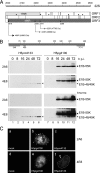
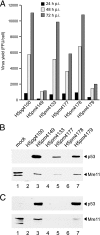
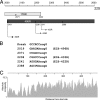
The adenovirus type 5 (Ad5) early region 1B 55-kDa (E1B-55K) protein is a multifunctional regulator of cell-cycle-independent virus replication that participates in many processes required for maximal virus production. As part of a study of E1B-55K function, we generated the Ad5 mutant H5pm4133, carrying stop codons after the second and seventh codons of the E1B reading frame, thereby eliminating synthesis of the full-length 55K product and its smaller derivatives. Unexpectedly, phenotypic studies revealed that H5pm4133 fully exhibits the characteristics of wild-type (wt) Ad5 in all assays tested. Immunoblot analyses demonstrated that H5pm4133 and wt Ad5 produce very low levels of two distinct polypeptides in the 48- to 49-kDa range, which lack the amino-terminal region but contain segments from the central and carboxy-terminal part of the 55K protein. Genetic and biochemical studies with different Ad5 mutants show that at least one of these isoforms consists of two closely migrating polypeptides of 433 amino acid residues (433R) and 422R, which are produced by translation initiation at two downstream AUG codons of the 55K reading frame. Significantly, a virus mutant producing low levels of the 433R isoform alone replicated to levels comparable to those of wt Ad5, demonstrating that this polypeptide provides essentially all functions of E1B-55K required to promote maximal virus growth in human tumor cells. Altogether, these results extend previous findings that the wt Ad5 E1B region encodes a series of smaller isoforms of E1B-55K and demonstrate that very low levels of at least one of these novel proteins (E1B-433R) are sufficient for a productive infection.
Figures




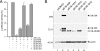
Similar articles
-
Diverse roles for E4orf3 at late times of infection revealed in an E1B 55-kilodalton protein mutant background.J Virol. 2004 Sep;78(18):9924-35. doi: 10.1128/JVI.78.18.9924-9935.2004. J Virol. 2004. PMID: 15331726 Free PMC article.
-
The replicative capacities of large E1B-null group A and group C adenoviruses are independent of host cell p53 status.J Virol. 1999 Mar;73(3):2074-83. doi: 10.1128/JVI.73.3.2074-2083.1999. J Virol. 1999. PMID: 9971789 Free PMC article.
-
Analyses of single-amino-acid substitution mutants of adenovirus type 5 E1B-55K protein.J Virol. 2001 May;75(9):4297-307. doi: 10.1128/JVI.75.9.4297-4307.2001. J Virol. 2001. PMID: 11287579 Free PMC article.
-
The adenovirus E1B 55-kilodalton and E4 open reading frame 6 proteins limit phosphorylation of eIF2alpha during the late phase of infection.J Virol. 2009 Oct;83(19):9970-82. doi: 10.1128/JVI.01113-09. Epub 2009 Jul 15. J Virol. 2009. PMID: 19605483 Free PMC article.
-
The biology of the adenovirus E1B 55K protein.FEBS Lett. 2019 Dec;593(24):3504-3517. doi: 10.1002/1873-3468.13694. Epub 2019 Dec 8. FEBS Lett. 2019. PMID: 31769868 Review.
Cited by
-
Adenovirus replaces mitotic checkpoint controls.J Virol. 2015 May;89(9):5083-96. doi: 10.1128/JVI.00213-15. Epub 2015 Feb 18. J Virol. 2015. PMID: 25694601 Free PMC article.
-
Adenovirus E1B-55K controls SUMO-dependent degradation of antiviral cellular restriction factors.J Virol. 2023 Nov 30;97(11):e0079123. doi: 10.1128/jvi.00791-23. Epub 2023 Nov 2. J Virol. 2023. PMID: 37916833 Free PMC article.
-
The Human Adenovirus Type 5 E4orf6/E1B55K E3 Ubiquitin Ligase Complex Can Mimic E1A Effects on E2F.mSphere. 2015 Nov 11;1(1):e00014-15. doi: 10.1128/mSphere.00014-15. eCollection 2016 Jan-Feb. mSphere. 2015. PMID: 27303679 Free PMC article.
-
Functional cooperation between human adenovirus type 5 early region 4, open reading frame 6 protein, and cellular homeobox protein HoxB7.J Virol. 2012 Aug;86(15):8296-308. doi: 10.1128/JVI.00222-12. Epub 2012 May 2. J Virol. 2012. PMID: 22553335 Free PMC article.
-
Transcriptional activation of the adenoviral genome is mediated by capsid protein VI.PLoS Pathog. 2012 Feb;8(2):e1002549. doi: 10.1371/journal.ppat.1002549. Epub 2012 Feb 23. PLoS Pathog. 2012. PMID: 22427750 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

