CDK9 directs H2B monoubiquitination and controls replication-dependent histone mRNA 3'-end processing
- PMID: 19575011
- PMCID: PMC2726677
- DOI: 10.1038/embor.2009.108
CDK9 directs H2B monoubiquitination and controls replication-dependent histone mRNA 3'-end processing
Abstract
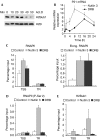
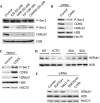
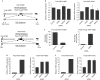
Post-translational histone modifications have essential roles in controlling nuclear processes; however, the specific mechanisms regulating these modifications and their combinatorial activities remain elusive. Cyclin-dependent kinase 9 (CDK9) regulates gene expression by phosphorylating transcriptional regulatory proteins, including the RNA polymerase II carboxy-terminal domain. Here, we show that CDK9 activity is essential for maintaining global and gene-associated levels of histone H2B monoubiquitination (H2Bub1). Furthermore, CDK9 activity and H2Bub1 help to maintain correct replication-dependent histone messenger RNA (mRNA) 3'-end processing. CDK9 knockdown consistently resulted in inefficient recognition of the correct mRNA 3'-end cleavage site and led to increased read-through of RNA polymerase II to an alternative downstream polyadenylation signal. Thus, CDK9 acts to integrate phosphorylation during transcription with chromatin modifications to control co-transcriptional histone mRNA processing.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
Phosphorylation by cyclin-dependent kinase-9 controls ubiquitin-conjugating enzyme-2A function.Cell Cycle. 2012 Jun 1;11(11):2122-7. doi: 10.4161/cc.20548. Epub 2012 Jun 1. Cell Cycle. 2012. PMID: 22592529
-
A positive feedback loop links opposing functions of P-TEFb/Cdk9 and histone H2B ubiquitylation to regulate transcript elongation in fission yeast.PLoS Genet. 2012;8(8):e1002822. doi: 10.1371/journal.pgen.1002822. Epub 2012 Aug 2. PLoS Genet. 2012. PMID: 22876190 Free PMC article.
-
Induced G1 cell-cycle arrest controls replication-dependent histone mRNA 3' end processing through p21, NPAT and CDK9.Oncogene. 2010 May 13;29(19):2853-63. doi: 10.1038/onc.2010.42. Epub 2010 Mar 1. Oncogene. 2010. PMID: 20190802
-
Insights into the function of the human P-TEFb component CDK9 in the regulation of chromatin modifications and co-transcriptional mRNA processing.Cell Cycle. 2009 Nov 15;8(22):3636-42. doi: 10.4161/cc.8.22.9890. Epub 2009 Nov 24. Cell Cycle. 2009. PMID: 19844166 Review.
-
The enigmatic role of H2Bub1 in cancer.FEBS Lett. 2012 Jun 4;586(11):1592-601. doi: 10.1016/j.febslet.2012.04.002. Epub 2012 Apr 13. FEBS Lett. 2012. PMID: 22564770 Review.
Cited by
-
RNF40 exerts stage-dependent functions in differentiating osteoblasts and is essential for bone cell crosstalk.Cell Death Differ. 2021 Feb;28(2):700-714. doi: 10.1038/s41418-020-00614-w. Epub 2020 Sep 8. Cell Death Differ. 2021. PMID: 32901120 Free PMC article.
-
Cdk7: a kinase at the core of transcription and in the crosshairs of cancer drug discovery.Transcription. 2019 Apr;10(2):47-56. doi: 10.1080/21541264.2018.1553483. Epub 2018 Dec 6. Transcription. 2019. PMID: 30488763 Free PMC article. Review.
-
CDK9 inhibitors in acute myeloid leukemia.J Exp Clin Cancer Res. 2018 Feb 23;37(1):36. doi: 10.1186/s13046-018-0704-8. J Exp Clin Cancer Res. 2018. PMID: 29471852 Free PMC article. Review.
-
HEXIM1 plays a critical role in the inhibition of the androgen receptor by anti-androgens.Biochem J. 2014 Sep 1;462(2):315-27. doi: 10.1042/BJ20140174. Biochem J. 2014. PMID: 24844355 Free PMC article.
-
Ctr9, a key subunit of PAFc, affects global estrogen signaling and drives ERα-positive breast tumorigenesis.Genes Dev. 2015 Oct 15;29(20):2153-67. doi: 10.1101/gad.268722.115. Genes Dev. 2015. PMID: 26494790 Free PMC article.
References
-
- Berger SL (2007) The complex language of chromatin regulation during transcription. Nature 447: 407–412 - PubMed
-
- Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D (2007) Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science 318: 1780–1782 - PubMed
-
- Davie JR, Murphy LC (1990) Level of ubiquitinated histone H2B in chromatin is coupled to ongoing transcription. Biochemistry 29: 4752–4757 - PubMed
-
- Egloff S, Murphy S (2008) Cracking the RNA polymerase II CTD code. Trends Genet 24: 280–288 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

