Confined dynamics of a ribosome-bound nascent globin: Cone angle analysis of fluorescence depolarization decays in the presence of two local motions
- PMID: 19569194
- PMCID: PMC2786964
- DOI: 10.1002/pro.196
Confined dynamics of a ribosome-bound nascent globin: Cone angle analysis of fluorescence depolarization decays in the presence of two local motions
Erratum in
- Protein Sci. 2010 Aug;19(8):1600
Abstract
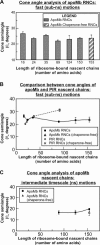
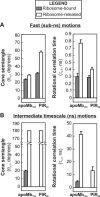
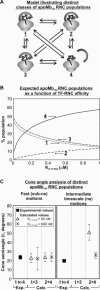
We still know very little about how proteins achieve their native three-dimensional structure in vitro and in the cell. Folding studies as proteins emerge from the mega Dalton-sized ribosome pose special challenges due to the large size and complicated nature of the ribosome-nascent chain complex. This work introduces a combination of three-component analysis of fluorescence depolarization decays (including the presence of two local motions) and in-cone analysis of diffusive local dynamics to investigate the spatial constraints experienced by a protein emerging from the ribosomal tunnel. We focus on E. coli ribosomes and an all-alpha-helical nascent globin in the presence and absence of the cotranslationally active chaperones DnaK and trigger factor. The data provide insights on the dynamic nature and structural plasticity of ribosome-nascent chain complexes. We find that the sub-ns motions of the N-terminal fluorophore, reporting on the globin dynamics in the vicinity of the N terminus, are highly constrained both inside and outside the ribosomal tunnel, resulting in high-order parameters (>0.85) and small cone semiangles (<30 degrees ). The shorter globin chains buried inside the tunnel are less spatially constrained than those of a reference sequence from a natively unfolded protein, suggesting either that the two nascent chain sequences have a different secondary structure and therefore sample different regions of the tunnel or that the tunnel undergoes local structural adjustments to accommodate the globin sequence. Longer globins emerging out of the ribosomal tunnel are also found to have highly spatially constrained slow (ns) motions. There are no observable spectroscopic changes in the absence of bound chaperones.
Figures
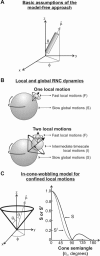
 ), defining the fluorophore's symmetry axis, is colinear with either the emission or excitation dipoles of the fluorophore and is allowed stochastic wobbling motions (described by the angle θ) within a static cone defined by the cone semiangle θo, in a reference frame attached to the macromolecule with the z axis representing the normal to the macromolecular surface. The motions are randomly distributed in the XY plane and therefore independent of the azimuthal angle φ. The plot on the right illustrates the dependence of the order parameter S and its square S2 on the cone semiangle θo.
), defining the fluorophore's symmetry axis, is colinear with either the emission or excitation dipoles of the fluorophore and is allowed stochastic wobbling motions (described by the angle θ) within a static cone defined by the cone semiangle θo, in a reference frame attached to the macromolecule with the z axis representing the normal to the macromolecular surface. The motions are randomly distributed in the XY plane and therefore independent of the azimuthal angle φ. The plot on the right illustrates the dependence of the order parameter S and its square S2 on the cone semiangle θo.

Similar articles
-
Dynamic fluorescence depolarization: a powerful tool to explore protein folding on the ribosome.Methods. 2010 Sep;52(1):57-73. doi: 10.1016/j.ymeth.2010.06.001. Epub 2010 Jun 8. Methods. 2010. PMID: 20685617 Free PMC article. Review.
-
Tethering creates unusual kinetics for ribosome-associated chaperones with nascent chains.Protein Pept Lett. 2009;16(6):631-4. doi: 10.2174/092986609788490195. Protein Pept Lett. 2009. PMID: 19519521
-
Chain dynamics of nascent polypeptides emerging from the ribosome.ACS Chem Biol. 2008 Sep 19;3(9):555-66. doi: 10.1021/cb800059u. Epub 2008 Aug 22. ACS Chem Biol. 2008. PMID: 18717565 Free PMC article.
-
Molecular mechanism and structure of Trigger Factor bound to the translating ribosome.EMBO J. 2008 Jun 4;27(11):1622-32. doi: 10.1038/emboj.2008.89. Epub 2008 May 22. EMBO J. 2008. PMID: 18497744 Free PMC article.
-
A cradle for new proteins: trigger factor at the ribosome.Curr Opin Struct Biol. 2005 Apr;15(2):204-12. doi: 10.1016/j.sbi.2005.03.005. Curr Opin Struct Biol. 2005. PMID: 15837180 Review.
Cited by
-
Autotransporters: The Cellular Environment Reshapes a Folding Mechanism to Promote Protein Transport.J Phys Chem Lett. 2012 Apr 2;3(8):1063-1071. doi: 10.1021/jz201654k. J Phys Chem Lett. 2012. PMID: 23687560 Free PMC article.
-
An intrinsically disordered nascent protein interacts with specific regions of the ribosomal surface near the exit tunnel.Commun Biol. 2021 Oct 29;4(1):1236. doi: 10.1038/s42003-021-02752-4. Commun Biol. 2021. PMID: 34716402 Free PMC article.
-
Folding up and Moving on-Nascent Protein Folding on the Ribosome.J Mol Biol. 2018 Oct 26;430(22):4580-4591. doi: 10.1016/j.jmb.2018.06.050. Epub 2018 Jul 5. J Mol Biol. 2018. PMID: 29981746 Free PMC article. Review.
-
Dynamic fluorescence depolarization: a powerful tool to explore protein folding on the ribosome.Methods. 2010 Sep;52(1):57-73. doi: 10.1016/j.ymeth.2010.06.001. Epub 2010 Jun 8. Methods. 2010. PMID: 20685617 Free PMC article. Review.
-
Revealing Conformational Variants of Solution-Phase Intrinsically Disordered Tau Protein at the Single-Molecule Level.Angew Chem Int Ed Engl. 2017 Dec 4;56(49):15584-15588. doi: 10.1002/anie.201708242. Epub 2017 Nov 14. Angew Chem Int Ed Engl. 2017. PMID: 29063723 Free PMC article.
References
-
- Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. - PubMed
-
- Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell (Cambridge, MA) 2001;107:679–688. - PubMed
-
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. - PubMed
-
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, and Cate JHD. Structures of the bacterial ribosome at 3.5 .ANG. resolution. Science. 2005;310:827–834. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

