alpha-Helical domains promote translocation of intrinsically disordered polypeptides into the endoplasmic reticulum
- PMID: 19561072
- PMCID: PMC2782031
- DOI: 10.1074/jbc.M109.023135
alpha-Helical domains promote translocation of intrinsically disordered polypeptides into the endoplasmic reticulum
Abstract
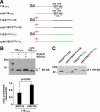
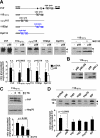
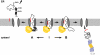
Co-translational import into the endoplasmic reticulum (ER) is primarily controlled by N-terminal signal sequences that mediate targeting of the ribosome-nascent chain complex to the Sec61/translocon and initiate the translocation process. Here we show that after targeting to the translocon the secondary structure of the nascent polypeptide chain can significantly modulate translocation efficiency. ER-targeted polypeptides dominated by unstructured domains failed to efficiently translocate into the ER lumen and were subjected to proteasomal degradation via a co-translocational/preemptive pathway. Productive ER import could be reinstated by increasing the amount of alpha-helical domains, whereas more effective ER signal sequences had only a minor effect on ER import efficiency of unstructured polypeptides. ER stress and overexpression of p58(IPK) promoted the co-translocational degradation pathway. Moreover polypeptides with unstructured domains at their N terminus were specifically targeted to proteasomal degradation under these conditions. Our study indicates that extended unstructured domains are signals to dispose ER-targeted proteins via a co-translocational, preemptive quality control pathway.
Figures



Similar articles
-
The Sec61/SecY complex is inherently deficient in translocating intrinsically disordered proteins.J Biol Chem. 2017 Dec 29;292(52):21383-21396. doi: 10.1074/jbc.M117.788067. Epub 2017 Oct 30. J Biol Chem. 2017. PMID: 29084847 Free PMC article.
-
The α-helical structure of prodomains promotes translocation of intrinsically disordered neuropeptide hormones into the endoplasmic reticulum.J Biol Chem. 2013 May 17;288(20):13961-13973. doi: 10.1074/jbc.M112.430264. Epub 2013 Mar 26. J Biol Chem. 2013. PMID: 23532840 Free PMC article.
-
Structural features within the nascent chain regulate alternative targeting of secretory proteins to mitochondria.EMBO J. 2013 Apr 3;32(7):1036-51. doi: 10.1038/emboj.2013.46. Epub 2013 Mar 12. EMBO J. 2013. PMID: 23481258 Free PMC article.
-
A clearer picture of the ER translocon complex.J Cell Sci. 2020 Feb 4;133(3):jcs231340. doi: 10.1242/jcs.231340. J Cell Sci. 2020. PMID: 32019826 Review.
-
Functions and Mechanisms of the Human Ribosome-Translocon Complex.Subcell Biochem. 2019;93:83-141. doi: 10.1007/978-3-030-28151-9_4. Subcell Biochem. 2019. PMID: 31939150 Review.
Cited by
-
Transgenic Overexpression of the Disordered Prion Protein N1 Fragment in Mice Does Not Protect Against Neurodegenerative Diseases Due to Impaired ER Translocation.Mol Neurobiol. 2020 Jun;57(6):2812-2829. doi: 10.1007/s12035-020-01917-2. Epub 2020 May 4. Mol Neurobiol. 2020. PMID: 32367491 Free PMC article.
-
Anti-prion drug mPPIg5 inhibits PrP(C) conversion to PrP(Sc).PLoS One. 2013;8(1):e55282. doi: 10.1371/journal.pone.0055282. Epub 2013 Jan 28. PLoS One. 2013. PMID: 23383136 Free PMC article.
-
The Sec61/SecY complex is inherently deficient in translocating intrinsically disordered proteins.J Biol Chem. 2017 Dec 29;292(52):21383-21396. doi: 10.1074/jbc.M117.788067. Epub 2017 Oct 30. J Biol Chem. 2017. PMID: 29084847 Free PMC article.
-
Cotranslational stabilization of Sec62/63 within the ER Sec61 translocon is controlled by distinct substrate-driven translocation events.Mol Cell. 2015 Apr 16;58(2):269-83. doi: 10.1016/j.molcel.2015.02.018. Epub 2015 Mar 19. Mol Cell. 2015. PMID: 25801167 Free PMC article.
-
VCP/p97 mediates nuclear targeting of non-ER-imported prion protein to maintain proteostasis.Life Sci Alliance. 2024 Apr 3;7(6):e202302456. doi: 10.26508/lsa.202302456. Print 2024 Jun. Life Sci Alliance. 2024. PMID: 38570188 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

