Fluorescence-based assay for phenotypic characterization of human cytomegalovirus polymerase mutations regarding drug susceptibility and viral replicative fitness
- PMID: 19546362
- PMCID: PMC2737853
- DOI: 10.1128/AAC.00165-09
Fluorescence-based assay for phenotypic characterization of human cytomegalovirus polymerase mutations regarding drug susceptibility and viral replicative fitness
Abstract
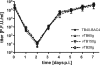
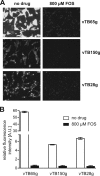
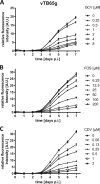
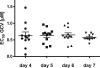
One essential prerequisite for genotypic drug susceptibility testing of human cytomegalovirus (HCMV) is the phenotypic characterization of mutations identified in the viral protein kinase gene UL97 and the viral DNA polymerase gene UL54 regarding their quantitative impact on drug susceptibility. We developed a new method for phenotypic characterization of UL54 mutations with regard to polymerase activity, viral replication, and drug susceptibility. To determine the most suitable viral indicator gene, enhanced green fluorescence protein was C-terminally fused to the HCMV early-late protein UL83 (pp65) or the late proteins UL32 (pp150) and UL99 (pp28), resulting in reporter viruses vTB65g, vTB150g, and vTB28g. vTB65g proved to be superior to the other constructs due to its favorable signal-to-noise ratio and was therefore used to establish the optimum conditions for our assay. The UL54 E756K and D413E mutations were introduced into vTB65g by markerless bacterial artificial chromosome mutagenesis, resulting in virus strains vE756Kg and vD413Eg. The drug susceptibility phenotypes of vE756Kg and vD413Eg were comparable to those previously reported. Furthermore, we found a reduced replicative fitness of vE756Kg by measuring fluorescence intensity as well as by conventional virus growth kinetics. Decreased fluorescence signals of vE756Kg- and vD413Eg-infected cells at late times of infection suggested a reduced polymerase activity, which was confirmed by real-time PCR quantification of the newly synthesized viral DNAs. This new fluorescence-based assay is a highly reproducible method for the phenotypic characterization of mutations potentially influencing drug susceptibility, viral replicative fitness, and polymerase activity of HCMV after marker transfer.
Figures
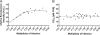
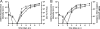

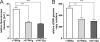
Similar articles
-
Identification of newly detected, drug-related HCMV UL97- and UL54-mutations using a modified plaque reduction assay.J Clin Virol. 2015 Aug;69:150-5. doi: 10.1016/j.jcv.2015.06.090. Epub 2015 Jun 19. J Clin Virol. 2015. PMID: 26209398
-
Identification of resistance-associated HCMV UL97- and UL54-mutations and a UL97-polymporphism with impact on phenotypic drug-resistance.Antiviral Res. 2016 Jul;131:1-8. doi: 10.1016/j.antiviral.2016.04.002. Epub 2016 Apr 4. Antiviral Res. 2016. PMID: 27058773
-
Human cytomegalovirus replication is strictly inhibited by siRNAs targeting UL54, UL97 or UL122/123 gene transcripts.PLoS One. 2014 Jun 2;9(6):e97231. doi: 10.1371/journal.pone.0097231. eCollection 2014. PLoS One. 2014. PMID: 24887060 Free PMC article.
-
Clinical and biologic aspects of human cytomegalovirus resistance to antiviral drugs.Hum Immunol. 2004 May;65(5):403-9. doi: 10.1016/j.humimm.2004.02.007. Hum Immunol. 2004. PMID: 15172438 Review.
-
[Analysis of ganciclovir-resistant cytomegalovirus].Nihon Rinsho. 1998 Jan;56(1):151-5. Nihon Rinsho. 1998. PMID: 9465681 Review. Japanese.
Cited by
-
An MHV-68 Mutator Phenotype Mutant Virus, Confirmed by CRISPR/Cas9-Mediated Gene Editing of the Viral DNA Polymerase Gene, Shows Reduced Viral Fitness.Viruses. 2021 May 26;13(6):985. doi: 10.3390/v13060985. Viruses. 2021. PMID: 34073189 Free PMC article.
-
Dynamic monitoring of viral gene expression reveals rapid antiviral effects of CD8 T cells recognizing the HCMV-pp65 antigen.Front Immunol. 2024 Jul 15;15:1439184. doi: 10.3389/fimmu.2024.1439184. eCollection 2024. Front Immunol. 2024. PMID: 39104541 Free PMC article.
-
A Luciferase Gene Driven by an Alphaherpesviral Promoter Also Responds to Immediate Early Antigens of the Betaherpesvirus HCMV, Allowing Comparative Analyses of Different Human Herpesviruses in One Reporter Cell Line.PLoS One. 2017 Jan 6;12(1):e0169580. doi: 10.1371/journal.pone.0169580. eCollection 2017. PLoS One. 2017. PMID: 28060895 Free PMC article.
-
Drug Susceptibility and Replicative Capacity of Multidrug-Resistant Recombinant Human Cytomegalovirus Harboring Mutations in UL56 and UL54 Genes.Antimicrob Agents Chemother. 2017 Oct 24;61(11):e01044-17. doi: 10.1128/AAC.01044-17. Print 2017 Nov. Antimicrob Agents Chemother. 2017. PMID: 28807919 Free PMC article.
-
Novel method based on "en passant" mutagenesis coupled with a gaussia luciferase reporter assay for studying the combined effects of human cytomegalovirus mutations.J Clin Microbiol. 2013 Oct;51(10):3216-24. doi: 10.1128/JCM.01275-13. Epub 2013 Jul 17. J Clin Microbiol. 2013. PMID: 23863570 Free PMC article.
References
-
- Avery, R. K. 2007. Management of late, recurrent, and resistant cytomegalovirus in transplant patients. Transpl. Rev. 21:65-76.
-
- Baldanti, F., N. Lurain, and G. Gerna. 2004. Clinical and biologic aspects of human cytomegalovirus resistance to antiviral drugs. Hum. Immunol. 65:403-409. - PubMed
-
- Biron, K. K. 2006. Antiviral drugs for cytomegalovirus diseases. Antivir. Res. 71:154-163. - PubMed
-
- Chevillotte, M., S. Landwehr, L. Linta, G. Frascaroli, A. Luske, C. Buser, T. Mertens, and J. von Einem. 2009. Major tegument protein pp65 of human cytomegalovirus is required for the incorporation of pUL69 and pUL97 into the virus particle and for viral growth in macrophages. J. Virol. 83:2480-2490. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

