Liver sinusoidal endothelial cells are a site of murine cytomegalovirus latency and reactivation
- PMID: 19535440
- PMCID: PMC2738169
- DOI: 10.1128/JVI.00870-09
Liver sinusoidal endothelial cells are a site of murine cytomegalovirus latency and reactivation
Abstract
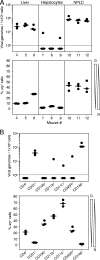
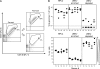
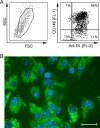
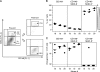
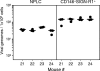
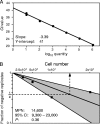
Latent cytomegalovirus (CMV) is frequently transmitted by organ transplantation, and its reactivation under conditions of immunosuppressive prophylaxis against graft rejection by host-versus-graft disease bears a risk of graft failure due to viral pathogenesis. CMV is the most common cause of infection following liver transplantation. Although hematopoietic cells of the myeloid lineage are a recognized source of latent CMV, the cellular sites of latency in the liver are not comprehensively typed. Here we have used the BALB/c mouse model of murine CMV infection to identify latently infected hepatic cell types. We performed sex-mismatched bone marrow transplantation with male donors and female recipients to generate latently infected sex chromosome chimeras, allowing us to distinguish between Y-chromosome (gene sry or tdy)-positive donor-derived hematopoietic descendants and Y-chromosome-negative cells of recipients' tissues. The viral genome was found to localize primarily to sry-negative CD11b(-) CD11c(-) CD31(+) CD146(+) cells lacking major histocompatibility complex class II antigen (MHC-II) but expressing murine L-SIGN. This cell surface phenotype is typical of liver sinusoidal endothelial cells (LSECs). Notably, sry-positive CD146(+) cells were distinguished by the expression of MHC-II and did not harbor latent viral DNA. In this model, the frequency of latently infected cells was found to be 1 to 2 per 10(4) LSECs, with an average copy number of 9 (range, 4 to 17) viral genomes. Ex vivo-isolated, latently infected LSECs expressed the viral genes m123/ie1 and M122/ie3 but not M112-M113/e1, M55/gB, or M86/MCP. Importantly, in an LSEC transfer model, infectious virus reactivated from recipients' tissue explants with an incidence of one reactivation per 1,000 viral-genome-carrying LSECs. These findings identified LSECs as the main cellular site of murine CMV latency and reactivation in the liver.
Figures



Similar articles
-
Role for tumor necrosis factor alpha in murine cytomegalovirus transcriptional reactivation in latently infected lungs.J Virol. 2005 Jan;79(1):326-40. doi: 10.1128/JVI.79.1.326-340.2005. J Virol. 2005. PMID: 15596827 Free PMC article.
-
Murine cytomegalovirus immediate-early 1 gene expression correlates with increased GVHD after allogeneic hematopoietic cell transplantation in recipients reactivating from latent infection.PLoS One. 2013 Apr 15;8(4):e61841. doi: 10.1371/journal.pone.0061841. Print 2013. PLoS One. 2013. PMID: 23596528 Free PMC article.
-
Stochastic Episodes of Latent Cytomegalovirus Transcription Drive CD8 T-Cell "Memory Inflation" and Avoid Immune Evasion.Front Immunol. 2021 Apr 22;12:668885. doi: 10.3389/fimmu.2021.668885. eCollection 2021. Front Immunol. 2021. PMID: 33968074 Free PMC article.
-
Mouse models of cytomegalovirus latency: overview.J Clin Virol. 2002 Aug;25 Suppl 2:S23-36. doi: 10.1016/s1386-6532(02)00087-2. J Clin Virol. 2002. PMID: 12361754 Review.
-
Cellular reservoirs of latent cytomegaloviruses.Med Microbiol Immunol. 2019 Aug;208(3-4):391-403. doi: 10.1007/s00430-019-00592-y. Epub 2019 Apr 22. Med Microbiol Immunol. 2019. PMID: 31011793 Review.
Cited by
-
Investigating the Dynamics of MCMV-Specific CD8+ T Cell Responses in Individual Hosts.Front Immunol. 2019 Jun 19;10:1358. doi: 10.3389/fimmu.2019.01358. eCollection 2019. Front Immunol. 2019. PMID: 31281313 Free PMC article.
-
Immunotherapy of cytomegalovirus infection by low-dose adoptive transfer of antiviral CD8 T cells relies on substantial post-transfer expansion of central memory cells but not effector-memory cells.PLoS Pathog. 2023 Nov 16;19(11):e1011643. doi: 10.1371/journal.ppat.1011643. eCollection 2023 Nov. PLoS Pathog. 2023. PMID: 37972198 Free PMC article.
-
Analysis of time-course gene expression profiles of sinusoidal endothelial cells during liver regeneration in rats.Mol Cell Biochem. 2011 Apr;350(1-2):215-27. doi: 10.1007/s11010-010-0701-5. Epub 2011 Jan 8. Mol Cell Biochem. 2011. PMID: 21221724
-
Efficient uptake of blood-borne BK and JC polyomavirus-like particles in endothelial cells of liver sinusoids and renal vasa recta.PLoS One. 2014 Nov 6;9(11):e111762. doi: 10.1371/journal.pone.0111762. eCollection 2014. PLoS One. 2014. PMID: 25375646 Free PMC article.
-
Cytomegalovirus Hepatitis in Immunocompetent and Immunocompromised Hosts.J Clin Transl Hepatol. 2021 Feb 28;9(1):106-115. doi: 10.14218/JCTH.2020.00088. Epub 2021 Jan 4. J Clin Transl Hepatol. 2021. PMID: 33604261 Free PMC article. Review.
References
-
- Agresti, A. 1992. A survey of exact inference for contingency tables. Stat. Sci. 7131-177.
-
- Alterio de Goss, M., R. Holtappels, H.-P. Steffens, J. Podlech, P. Angele, L. Dreher, D. Thomas, and M. J. Reddehase. 1998. Control of cytomegalovirus in bone marrow transplantation chimeras lacking the prevailing antigen-presenting molecule in recipient tissues rests primarily on recipient-derived CD8 T cells. J. Virol. 727733-7744. - PMC - PubMed
-
- Ambion. 2009. MEGAscript™ high yield transcription kit instruction manual (manual version 0209). Ambion, Austin, TX.
-
- Asahara, T., H. Masuda, T. Takahashi, C. Kalka, C. Pastore, M. Silver, M. Kearne, M. Magner, and J. M. Isner. 1999. Bone marrow origin of endothelial progenitor cells responsible for postnatal vasculogenesis in physiological and pathological neovascularization. Circ. Res. 85221-228. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

