Crystal structures of human SIRT3 displaying substrate-induced conformational changes
- PMID: 19535340
- PMCID: PMC2782032
- DOI: 10.1074/jbc.M109.014928
Crystal structures of human SIRT3 displaying substrate-induced conformational changes
Abstract
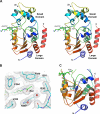
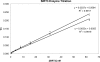
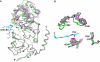
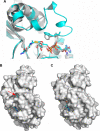
SIRT3 is a major mitochondrial NAD(+)-dependent protein deacetylase playing important roles in regulating mitochondrial metabolism and energy production and has been linked to the beneficial effects of exercise and caloric restriction. SIRT3 is emerging as a potential therapeutic target to treat metabolic and neurological diseases. We report the first sets of crystal structures of human SIRT3, an apo-structure with no substrate, a structure with a peptide containing acetyl lysine of its natural substrate acetyl-CoA synthetase 2, a reaction intermediate structure trapped by a thioacetyl peptide, and a structure with the dethioacetylated peptide bound. These structures provide insights into the conformational changes induced by the two substrates required for the reaction, the acetylated substrate peptide and NAD(+). In addition, the binding study by isothermal titration calorimetry suggests that the acetylated peptide is the first substrate to bind to SIRT3, before NAD(+). These structures and biophysical studies provide key insight into the structural and functional relationship of the SIRT3 deacetylation activity.
Figures



Similar articles
-
Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2.Proc Natl Acad Sci U S A. 2006 Jul 5;103(27):10224-10229. doi: 10.1073/pnas.0603968103. Epub 2006 Jun 20. Proc Natl Acad Sci U S A. 2006. PMID: 16788062 Free PMC article.
-
Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5.J Mol Biol. 2008 Oct 10;382(3):790-801. doi: 10.1016/j.jmb.2008.07.048. Epub 2008 Jul 25. J Mol Biol. 2008. PMID: 18680753
-
SIRT3 substrate specificity determined by peptide arrays and machine learning.ACS Chem Biol. 2011 Feb 18;6(2):146-57. doi: 10.1021/cb100218d. Epub 2010 Nov 1. ACS Chem Biol. 2011. PMID: 20945913 Free PMC article.
-
SIRT3 regulates mitochondrial protein acetylation and intermediary metabolism.Cold Spring Harb Symp Quant Biol. 2011;76:267-77. doi: 10.1101/sqb.2011.76.010850. Epub 2011 Nov 23. Cold Spring Harb Symp Quant Biol. 2011. PMID: 22114326 Review.
-
Mitochondrial sirtuins.Biochim Biophys Acta. 2010 Aug;1804(8):1645-51. doi: 10.1016/j.bbapap.2009.12.021. Epub 2010 Jan 7. Biochim Biophys Acta. 2010. PMID: 20060508 Review.
Cited by
-
Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome.Mol Cell. 2013 Jan 10;49(1):186-99. doi: 10.1016/j.molcel.2012.10.024. Epub 2012 Nov 29. Mol Cell. 2013. PMID: 23201123 Free PMC article.
-
Functions of mammalian SIRT4 in cellular metabolism and research progress in human cancer.Oncol Lett. 2020 Oct;20(4):11. doi: 10.3892/ol.2020.11872. Epub 2020 Jul 15. Oncol Lett. 2020. PMID: 32774484 Free PMC article. Review.
-
Sirt5 deacylation activities show differential sensitivities to nicotinamide inhibition.PLoS One. 2012;7(9):e45098. doi: 10.1371/journal.pone.0045098. Epub 2012 Sep 19. PLoS One. 2012. PMID: 23028781 Free PMC article.
-
Efficient demyristoylase activity of SIRT2 revealed by kinetic and structural studies.Sci Rep. 2015 Feb 23;5:8529. doi: 10.1038/srep08529. Sci Rep. 2015. PMID: 25704306 Free PMC article.
-
Novel Thiazole-Based SIRT2 Inhibitors Discovered via Molecular Modelling Studies and Enzymatic Assays.Pharmaceuticals (Basel). 2023 Sep 18;16(9):1316. doi: 10.3390/ph16091316. Pharmaceuticals (Basel). 2023. PMID: 37765125 Free PMC article.
References
-
- Dutnall R. N., Pillus L. (2001) Cell 105, 161–164 - PubMed
-
- Vaquero A., Scher M., Lee D., Erdjument-Bromage H., Tempst P., Reinberg D. (2004) Mol. Cell 16, 93–105 - PubMed
-
- Vaziri H., Dessain S. K., Ng Eaton E., Imai S. I., Frye R. A., Pandita T. K., Guarente L., Weinberg R. A. (2001) Cell 107, 149–159 - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

