Flipping of alkylated DNA damage bridges base and nucleotide excision repair
- PMID: 19516334
- PMCID: PMC2729916
- DOI: 10.1038/nature08076
Flipping of alkylated DNA damage bridges base and nucleotide excision repair
Abstract
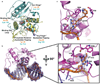
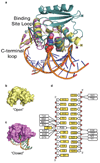
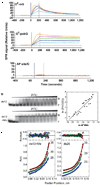
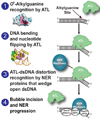
Alkyltransferase-like proteins (ATLs) share functional motifs with the cancer chemotherapy target O(6)-alkylguanine-DNA alkyltransferase (AGT) and paradoxically protect cells from the biological effects of DNA alkylation damage, despite lacking the reactive cysteine and alkyltransferase activity of AGT. Here we determine Schizosaccharomyces pombe ATL structures without and with damaged DNA containing the endogenous lesion O(6)-methylguanine or cigarette-smoke-derived O(6)-4-(3-pyridyl)-4-oxobutylguanine. These results reveal non-enzymatic DNA nucleotide flipping plus increased DNA distortion and binding pocket size compared to AGT. Our analysis of lesion-binding site conservation identifies new ATLs in sea anemone and ancestral archaea, indicating that ATL interactions are ancestral to present-day repair pathways in all domains of life. Genetic connections to mammalian XPG (also known as ERCC5) and ERCC1 in S. pombe homologues Rad13 and Swi10 and biochemical interactions with Escherichia coli UvrA and UvrC combined with structural results reveal that ATLs sculpt alkylated DNA to create a genetic and structural intersection of base damage processing with nucleotide excision repair.
Figures

Similar articles
-
Structural basis of O6-alkylguanine recognition by a bacterial alkyltransferase-like DNA repair protein.J Biol Chem. 2010 Apr 30;285(18):13736-41. doi: 10.1074/jbc.M109.093591. Epub 2010 Mar 8. J Biol Chem. 2010. PMID: 20212037 Free PMC article.
-
Alkyltransferase-like protein clusters scan DNA rapidly over long distances and recruit NER to alkyl-DNA lesions.Proc Natl Acad Sci U S A. 2020 Apr 28;117(17):9318-9328. doi: 10.1073/pnas.1916860117. Epub 2020 Apr 9. Proc Natl Acad Sci U S A. 2020. PMID: 32273391 Free PMC article.
-
Conserved structural motifs governing the stoichiometric repair of alkylated DNA by O(6)-alkylguanine-DNA alkyltransferase.Mutat Res. 2000 Aug 30;460(3-4):151-63. doi: 10.1016/s0921-8777(00)00024-0. Mutat Res. 2000. PMID: 10946226 Review.
-
Alkyltransferase-like protein (Atl1) distinguishes alkylated guanines for DNA repair using cation-π interactions.Proc Natl Acad Sci U S A. 2012 Nov 13;109(46):18755-60. doi: 10.1073/pnas.1209451109. Epub 2012 Oct 29. Proc Natl Acad Sci U S A. 2012. PMID: 23112169 Free PMC article.
-
Alkyltransferase-like proteins: molecular switches between DNA repair pathways.Cell Mol Life Sci. 2010 Nov;67(22):3749-62. doi: 10.1007/s00018-010-0405-8. Epub 2010 May 26. Cell Mol Life Sci. 2010. PMID: 20502938 Free PMC article. Review.
Cited by
-
Transcriptional mutagenesis dramatically alters genome-wide p53 transactivation landscape.Sci Rep. 2020 Aug 11;10(1):13513. doi: 10.1038/s41598-020-70412-4. Sci Rep. 2020. PMID: 32782319 Free PMC article.
-
X-ray scattering reveals disordered linkers and dynamic interfaces in complexes and mechanisms for DNA double-strand break repair impacting cell and cancer biology.Protein Sci. 2021 Sep;30(9):1735-1756. doi: 10.1002/pro.4133. Epub 2021 Jun 5. Protein Sci. 2021. PMID: 34056803 Free PMC article. Review.
-
Molecular coupling of DNA methylation and histone methylation.Epigenomics. 2010 Oct;2(5):657-69. doi: 10.2217/epi.10.44. Epigenomics. 2010. PMID: 21339843 Free PMC article. Review.
-
A new structural framework for integrating replication protein A into DNA processing machinery.Nucleic Acids Res. 2013 Feb 1;41(4):2313-27. doi: 10.1093/nar/gks1332. Epub 2013 Jan 8. Nucleic Acids Res. 2013. PMID: 23303776 Free PMC article.
-
Cooperative cluster formation, DNA bending and base-flipping by O6-alkylguanine-DNA alkyltransferase.Nucleic Acids Res. 2012 Sep 1;40(17):8296-308. doi: 10.1093/nar/gks574. Epub 2012 Jun 22. Nucleic Acids Res. 2012. PMID: 22730295 Free PMC article.
References
-
- Pegg AE. Repair of O6-alkylguanine by alkyltransferases. Mutat. Res. 2000;462:83–100. - PubMed
-
- Pauly GT, Hughes SH, Moschel RC. Comparison of mutagenesis by O6-methyl-and O6-ethylguanine and O4-methylthymine in Escherichia coli using double-stranded and gapped plasmids. Carcinogenesis. 1998;19:457–461. - PubMed
-
- Margison GP, Santibáñez-Koref MF. O6-Alkylguanine-DNA alkyltransferase: role in carcinogenesis and chemotherapy. Bioessays. 2002;24:255–266. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
- R01 CA059887-12/CA/NCI NIH HHS/United States
- R01 CA059887-13/CA/NCI NIH HHS/United States
- R01 CA059887/CA/NCI NIH HHS/United States
- R01 GM070662-05/GM/NIGMS NIH HHS/United States
- CA018137/CA/NCI NIH HHS/United States
- R01 GM070662-03/GM/NIGMS NIH HHS/United States
- CRUK_/Cancer Research UK/United Kingdom
- GM070662/GM/NIGMS NIH HHS/United States
- CA59887/CA/NCI NIH HHS/United States
- R01 GM070662-04/GM/NIGMS NIH HHS/United States
- R01 CA097209/CA/NCI NIH HHS/United States
- R01 GM070662-02/GM/NIGMS NIH HHS/United States
- R01 GM070662-01/GM/NIGMS NIH HHS/United States
- CA097209/CA/NCI NIH HHS/United States
- R01 GM070662/GM/NIGMS NIH HHS/United States
- R01 GM070662-06/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

