Structure of a mitochondrial ribosome with minimal RNA
- PMID: 19497863
- PMCID: PMC2700991
- DOI: 10.1073/pnas.0901631106
Structure of a mitochondrial ribosome with minimal RNA
Abstract
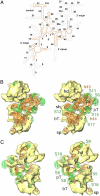
The Leishmania tarentolae mitochondrial ribosome (Lmr) is a minimal ribosomal RNA (rRNA)-containing ribosome. We have obtained a cryo-EM map of the Lmr. The map reveals several features that have not been seen in previously-determined structures of eubacterial or eukaryotic (cytoplasmic or organellar) ribosomes to our knowledge. Comparisons of the Lmr map with X-ray crystallographic and cryo-EM maps of the eubacterial ribosomes and a cryo-EM map of the mammalian mitochondrial ribosome show that (i) the overall structure of the Lmr is considerably more porous, (ii) the topology of the intersubunit space is significantly different, with fewer intersubunit bridges, but more tunnels, and (iii) several of the functionally-important rRNA regions, including the alpha-sarcin-ricin loop, have different relative positions within the structure. Furthermore, the major portions of the mRNA channel, the tRNA passage, and the nascent polypeptide exit tunnel contain Lmr-specific proteins, suggesting that the mechanisms for mRNA recruitment, tRNA interaction, and exiting of the nascent polypeptide in Lmr must differ markedly from the mechanisms deduced for ribosomes in other organisms. Our study identifies certain structural features that are characteristic solely of mitochondrial ribosomes and other features that are characteristic of both mitochondrial and chloroplast ribosomes (i.e., organellar ribosomes).
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins.Cell. 2003 Oct 3;115(1):97-108. doi: 10.1016/s0092-8674(03)00762-1. Cell. 2003. PMID: 14532006
-
Cryo-EM structure of the small subunit of the mammalian mitochondrial ribosome.Proc Natl Acad Sci U S A. 2014 May 20;111(20):7284-9. doi: 10.1073/pnas.1401657111. Epub 2014 May 5. Proc Natl Acad Sci U S A. 2014. PMID: 24799711 Free PMC article.
-
Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation.EMBO J. 2004 Mar 10;23(5):1008-19. doi: 10.1038/sj.emboj.7600102. Epub 2004 Feb 19. EMBO J. 2004. PMID: 14976550 Free PMC article.
-
The 55S mammalian mitochondrial ribosome and its tRNA-exit region.Biochimie. 2015 Jul;114:119-26. doi: 10.1016/j.biochi.2015.03.013. Epub 2015 Mar 20. Biochimie. 2015. PMID: 25797916 Free PMC article. Review.
-
Joachim Frank's Binding with the Ribosome.Structure. 2019 Mar 5;27(3):411-419. doi: 10.1016/j.str.2018.11.006. Epub 2018 Dec 27. Structure. 2019. PMID: 30595455 Free PMC article. Review.
Cited by
-
Mitochondrial evolution.Cold Spring Harb Perspect Biol. 2012 Sep 1;4(9):a011403. doi: 10.1101/cshperspect.a011403. Cold Spring Harb Perspect Biol. 2012. PMID: 22952398 Free PMC article. Review.
-
Proteins at the polypeptide tunnel exit of the yeast mitochondrial ribosome.J Biol Chem. 2010 Jun 18;285(25):19022-8. doi: 10.1074/jbc.M110.113837. Epub 2010 Apr 19. J Biol Chem. 2010. PMID: 20404317 Free PMC article.
-
Mitochondrial Neurodegenerative Diseases: Three Mitochondrial Ribosomal Proteins as Intermediate Stage in the Pathway That Associates Damaged Genes with Alzheimer's and Parkinson's.Biology (Basel). 2023 Jul 8;12(7):972. doi: 10.3390/biology12070972. Biology (Basel). 2023. PMID: 37508402 Free PMC article. Review.
-
Trans-splicing and RNA editing of LSU rRNA in Diplonema mitochondria.Nucleic Acids Res. 2014 Feb;42(4):2660-72. doi: 10.1093/nar/gkt1152. Epub 2013 Nov 19. Nucleic Acids Res. 2014. PMID: 24259427 Free PMC article.
-
The importance of the 45 S ribosomal small subunit-related complex for mitochondrial translation in Trypanosoma brucei.J Biol Chem. 2013 Nov 15;288(46):32963-78. doi: 10.1074/jbc.M113.501874. Epub 2013 Oct 2. J Biol Chem. 2013. PMID: 24089529 Free PMC article.
References
-
- Vickerman K, Preston TM. In: Biology of the Kinetoplastida. Lumsden WHR, Evans DA, editors. London: Academic; 1976. pp. 35–130.
-
- Lukeš J, Hashimi H, Zíková A. Unexplained complexity of the mitochondrial genome and transcriptome in kinetoplastid flagellates. Curr Genet. 2005;48:277–299. - PubMed
-
- Stuart KD, Schnaufer A, Ernst NL, Panigrahi A. Complex management: RNA editing in trypanosomes. Trends Biochem Sci. 2005;30:97–105. - PubMed
-
- Horváth A, Neboháčová M, Lukeš J, Maslov DA. Unusual polypeptide synthesis in the kinetoplast-mitochondria from Leishmania tarentolae. Identification of individual de novo translation products. J Biol Chem. 2002;277:7222–7230. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

