Diverse effects of individual mismatch repair components on transcription-induced CAG repeat instability in human cells
- PMID: 19497791
- PMCID: PMC3671857
- DOI: 10.1016/j.dnarep.2009.04.024
Diverse effects of individual mismatch repair components on transcription-induced CAG repeat instability in human cells
Abstract
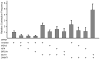
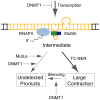
Several neurodegerative diseases are caused by expansion of a trinucleotide repeat tract in a critical gene. The mechanism of repeat instability is not yet defined, but in mice it requires MutSbeta, a complex of MSH2 and MSH3. We showed previously that transcription through a CAG repeat tract induces repeat instability in human cells via a pathway that requires the mismatch repair (MMR) components, MSH2 and MSH3, and the entire transcription-coupled nucleotide excision repair pathway [Y. Lin, V. Dion, J.H. Wilson, Transcription promotes contraction of CAG repeat tracts in human cells, Nat. Struct. Mol. Biol. 13 (2006) 179-180; Y. Lin, J.H. Wilson, Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair, Mol. Cell Biol. 27 (2007) 6209-6217]. Here, we examine the role of downstream MMR processing components on transcription-induced CAG instability, using our selection assay for repeat contraction. In contrast to knockdowns of MSH2 or MSH3, which reduce repeat contractions, we show that siRNA-mediated depletion of MLH1 or PMS2 increases contraction frequency. Knockdown of DNMT1, which has been identified as an MMR factor in genetic studies, also elevates the frequency of contraction. Simultaneous knockdowns of MLH1 or DNMT1 along with MSH2, XPA, or BRCA1, whose individual knockdowns each decrease CAG contraction, yield intermediate frequencies. In sharp contrast, double knockdown of MLH1 and DNMT1 additively increases the frequency of CAG contraction. These results show that MMR components can alter repeat stability in diverse ways, either enhancing or suppressing CAG contraction, and they provide insight into the influence of MMR components on transcription-induced CAG repeat instability.
Figures
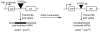
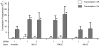

Similar articles
-
Mismatch repair enhances convergent transcription-induced cell death at trinucleotide repeats by activating ATR.DNA Repair (Amst). 2016 Jun;42:26-32. doi: 10.1016/j.dnarep.2016.03.016. Epub 2016 Apr 16. DNA Repair (Amst). 2016. PMID: 27131875 Free PMC article.
-
MutLα heterodimers modify the molecular phenotype of Friedreich ataxia.PLoS One. 2014 Jun 27;9(6):e100523. doi: 10.1371/journal.pone.0100523. eCollection 2014. PLoS One. 2014. PMID: 24971578 Free PMC article.
-
Differing patterns of genetic instability in mice deficient in the mismatch repair genes Pms2, Mlh1, Msh2, Msh3 and Msh6.Carcinogenesis. 2006 Dec;27(12):2402-8. doi: 10.1093/carcin/bgl079. Epub 2006 May 25. Carcinogenesis. 2006. PMID: 16728433 Free PMC article.
-
Disease-associated repeat instability and mismatch repair.DNA Repair (Amst). 2016 Feb;38:117-126. doi: 10.1016/j.dnarep.2015.11.008. Epub 2015 Dec 12. DNA Repair (Amst). 2016. PMID: 26774442 Review.
-
A review of mismatch repair gene transcripts: issues for interpretation of mRNA splicing assays.Clin Genet. 2015 Feb;87(2):100-8. doi: 10.1111/cge.12450. Epub 2014 Jul 26. Clin Genet. 2015. PMID: 24989436 Review.
Cited by
-
Antagonistic roles of canonical and Alternative-RPA in disease-associated tandem CAG repeat instability.Cell. 2023 Oct 26;186(22):4898-4919.e25. doi: 10.1016/j.cell.2023.09.008. Epub 2023 Oct 11. Cell. 2023. PMID: 37827155 Free PMC article.
-
Site-specific R-loops induce CGG repeat contraction and fragile X gene reactivation.Cell. 2023 Jun 8;186(12):2593-2609.e18. doi: 10.1016/j.cell.2023.04.035. Epub 2023 May 19. Cell. 2023. PMID: 37209683 Free PMC article.
-
Modifiers of CAG/CTG Repeat Instability: Insights from Mammalian Models.J Huntingtons Dis. 2021;10(1):123-148. doi: 10.3233/JHD-200426. J Huntingtons Dis. 2021. PMID: 33579861 Free PMC article. Review.
-
Mismatch repair enhances convergent transcription-induced cell death at trinucleotide repeats by activating ATR.DNA Repair (Amst). 2016 Jun;42:26-32. doi: 10.1016/j.dnarep.2016.03.016. Epub 2016 Apr 16. DNA Repair (Amst). 2016. PMID: 27131875 Free PMC article.
-
Fanconi anemia pathway regulates convergent transcription-induced cell death at trinucleotide repeats in human cells.Postdoc J. 2016 May;4(5):46-54. doi: 10.14304/surya.jpr.v4n5.1. Postdoc J. 2016. PMID: 27595121 Free PMC article.
References
-
- Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: mechanisms and common principles. Nat Rev Genet. 2005;6:743–755. - PubMed
-
- Pearson CE, Edamura KN, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet. 2005;6:729–742. - PubMed
-
- Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

