Rescue of key features of the p63-null epithelial phenotype by inactivation of Ink4a and Arf
- PMID: 19494829
- PMCID: PMC2711186
- DOI: 10.1038/emboj.2009.151
Rescue of key features of the p63-null epithelial phenotype by inactivation of Ink4a and Arf
Abstract
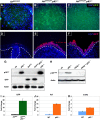
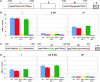
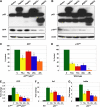
Mice lacking p63 cannot form skin, exhibit craniofacial and skeletal defects, and die soon after birth. The p63 gene regulates a complex network of target genes, and disruption of p63 has been shown to affect the maintenance of epithelial stem cells, the differentiation of keratinocytes, and the preservation of the adhesive properties of stratified epithelium. Here, we show that inactivation of p63 in mice is accompanied by aberrantly increased expression of the Ink4a and Arf tumour suppressor genes. In turn, anomalies of the p63-null mouse affecting the skin and skeleton are partially ameliorated in mice lacking either Ink4a or Arf. Rescue of epithelialization is accompanied by restoration of keratinocyte proliferative capacity both in vivo and in vitro and by expression of markers of squamous differentiation. Thus, in the absence of p63, abnormal upregulation of Ink4a and Arf is incompatible with skin development.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Similar articles
-
ΔNp63 knockout mice reveal its indispensable role as a master regulator of epithelial development and differentiation.Development. 2012 Feb;139(4):772-82. doi: 10.1242/dev.071191. Development. 2012. PMID: 22274697 Free PMC article.
-
Dynamic life of a skin keratinocyte: an intimate tryst with the master regulator p63.Indian J Exp Biol. 2011 Oct;49(10):721-31. Indian J Exp Biol. 2011. PMID: 22013738 Review.
-
Cited2 is an essential regulator of adult hematopoietic stem cells.Cell Stem Cell. 2009 Dec 4;5(6):659-65. doi: 10.1016/j.stem.2009.11.001. Cell Stem Cell. 2009. PMID: 19951693 Free PMC article.
-
Hdac1 and Hdac2 act redundantly to control p63 and p53 functions in epidermal progenitor cells.Dev Cell. 2010 Dec 14;19(6):807-18. doi: 10.1016/j.devcel.2010.10.015. Epub 2010 Nov 18. Dev Cell. 2010. PMID: 21093383 Free PMC article.
-
The regulation of INK4/ARF in cancer and aging.Cell. 2006 Oct 20;127(2):265-75. doi: 10.1016/j.cell.2006.10.003. Cell. 2006. PMID: 17055429 Review.
Cited by
-
Differential regulation of cutaneous oncoprotein HPVE6 by wtp53, mutant p53R248W and ΔNp63α is HPV type dependent.PLoS One. 2012;7(4):e35540. doi: 10.1371/journal.pone.0035540. Epub 2012 Apr 18. PLoS One. 2012. PMID: 22530045 Free PMC article.
-
TAp63: The fountain of youth.Aging (Albany NY). 2009 Oct 21;1(10):866-9. doi: 10.18632/aging.100095. Aging (Albany NY). 2009. PMID: 20157559 Free PMC article.
-
Induced multipotency in adult keratinocytes through down-regulation of ΔNp63 or DGCR8.Proc Natl Acad Sci U S A. 2014 Feb 4;111(5):E572-81. doi: 10.1073/pnas.1319743111. Epub 2014 Jan 21. Proc Natl Acad Sci U S A. 2014. PMID: 24449888 Free PMC article.
-
Dysregulated ΔNp63α inhibits expression of Ink4a/arf, blocks senescence, and promotes malignant conversion of keratinocytes.PLoS One. 2011;6(7):e21877. doi: 10.1371/journal.pone.0021877. Epub 2011 Jul 15. PLoS One. 2011. PMID: 21789189 Free PMC article.
-
Mutant p63 causes defective expansion of ectodermal progenitor cells and impaired FGF signalling in AEC syndrome.EMBO Mol Med. 2012 Mar;4(3):192-205. doi: 10.1002/emmm.201100199. Epub 2012 Jan 13. EMBO Mol Med. 2012. PMID: 22247000 Free PMC article.
References
-
- Akala OO, Park I-K, Qian D, Pihalja M, Becker MW, Clarke MF (2008) Long-term hematopoietic reconstitution by Trp53−/−, p16Inka−/−, p19Arf−/− multipotent progenitors. Nature 453: 228–232 - PubMed
-
- Bertwistle D, Zindy F, Sherr CJ, Roussel MF (2004) Monoclonal antibodies to the mouse p19(Arf) tumor suppressor protein. Hybrid Hybridomics 23: 293–300 - PubMed
-
- Bruggeman SW, Valk-Lingbeek ME, van der Stoop PP, Jacobs JJ, Kieboom K, Tanger E, Hulsman D, Leung C, Arsenijevic Y, Marino S, van Lohuizen M (2005) Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes Dev 19: 1438–1443 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

