Transcriptional competence of the integrated HIV-1 provirus at the nuclear periphery
- PMID: 19478796
- PMCID: PMC2726691
- DOI: 10.1038/emboj.2009.141
Transcriptional competence of the integrated HIV-1 provirus at the nuclear periphery
Abstract
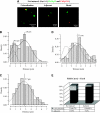
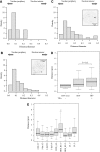
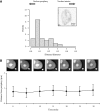
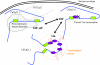
Spatial distribution of genes within the nucleus contributes to transcriptional control, allowing optimal gene expression as well as constitutive or regulated gene repression. Human immunodeficiency virus type 1 (HIV-1) integrates into host chromatin to transcribe and replicate its genome. Lymphocytes harbouring a quiescent but inducible provirus are a challenge to viral eradication in infected patients undergoing antiviral therapy. Therefore, our understanding of the contribution of sub-nuclear positioning to viral transcription may also have far-reaching implications in the pathology of the infection. To gain an insight into the conformation of chromatin at the site of HIV-1 integration, we investigated lymphocytes carrying a single latent provirus. In the silenced state, the provirus was consistently found at the nuclear periphery, associated in trans with a pericentromeric region of chromosome 12 in a significant number of quiescent cells. After induction of the transcription, this association was lost, although the location of the transcribing provirus remained peripheral. These results, extended to several other cell clones, unveil a novel mechanism of transcriptional silencing involved in HIV-1 post-transcriptional latency and reinforce the notion that gene transcription may also occur at the nuclear periphery.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Comment in
-
Gene activation at the edge of the nucleus.EMBO J. 2009 Aug 5;28(15):2145-6. doi: 10.1038/emboj.2009.148. EMBO J. 2009. PMID: 19654603 Free PMC article. No abstract available.
Similar articles
-
Nuclear positional control of HIV transcription in 4D.Nucleus. 2010 Jan-Feb;1(1):8-11. doi: 10.4161/nucl.1.1.10136. Nucleus. 2010. PMID: 21327098 Free PMC article.
-
HIV Provirus Stably Reproduces Parental Latent and Induced Transcription Phenotypes Regardless of the Chromosomal Integration Site.J Virol. 2016 May 12;90(11):5302-14. doi: 10.1128/JVI.02842-15. Print 2016 Jun 1. J Virol. 2016. PMID: 26984732 Free PMC article.
-
Proviruses with Long-Term Stable Expression Accumulate in Transcriptionally Active Chromatin Close to the Gene Regulatory Elements: Comparison of ASLV-, HIV- and MLV-Derived Vectors.Viruses. 2018 Mar 8;10(3):116. doi: 10.3390/v10030116. Viruses. 2018. PMID: 29517993 Free PMC article.
-
Factors controlling chromatin organization and nucleosome positioning for establishment and maintenance of HIV latency.Curr HIV Res. 2008 Jun;6(4):286-95. doi: 10.2174/157016208785132563. Curr HIV Res. 2008. PMID: 18691027 Review.
-
Attacking HIV provirus: therapeutic strategies to disrupt persistent infection.Infect Disord Drug Targets. 2006 Dec;6(4):369-76. doi: 10.2174/187152606779025824. Infect Disord Drug Targets. 2006. PMID: 17168802 Review.
Cited by
-
Targeting Epigenetics to Cure HIV-1: Lessons From (and for) Cancer Treatment.Front Cell Infect Microbiol. 2021 May 7;11:668637. doi: 10.3389/fcimb.2021.668637. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34026665 Free PMC article. Review.
-
Retroviral integration: Site matters: Mechanisms and consequences of retroviral integration site selection.Bioessays. 2015 Nov;37(11):1202-14. doi: 10.1002/bies.201500051. Epub 2015 Aug 21. Bioessays. 2015. PMID: 26293289 Free PMC article. Review.
-
Heterogeneity of HIV-1 latent reservoirs.Chin Med J (Engl). 2020 Dec 5;133(23):2867-2873. doi: 10.1097/CM9.0000000000001085. Chin Med J (Engl). 2020. PMID: 33273337 Free PMC article.
-
Dynamics of Viral and Host 3D Genome Structure upon Infection.J Microbiol Biotechnol. 2022 Dec 28;32(12):1515-1526. doi: 10.4014/jmb.2208.08020. Epub 2022 Sep 30. J Microbiol Biotechnol. 2022. PMID: 36398441 Free PMC article. Review.
-
Chromatin maturation of the HIV-1 provirus in primary resting CD4+ T cells.PLoS Pathog. 2020 Jan 30;16(1):e1008264. doi: 10.1371/journal.ppat.1008264. eCollection 2020 Jan. PLoS Pathog. 2020. PMID: 31999790 Free PMC article.
References
-
- Andrulis ED, Neiman AM, Zappulla DC, Sternglanz R (1998) Perinuclear localization of chromatin facilitates transcriptional silencing. Nature 394: 592–595 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

