Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells
- PMID: 19477429
- PMCID: PMC2693960
- DOI: 10.1016/j.ccr.2009.03.018
Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells
Abstract
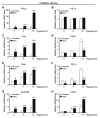
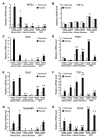
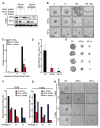
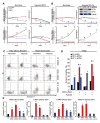
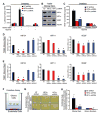
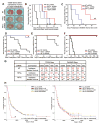
Glioblastomas are lethal cancers characterized by florid angiogenesis promoted in part by glioma stem cells (GSCs). Because hypoxia regulates angiogenesis, we examined hypoxic responses in GSCs. We now demonstrate that hypoxia-inducible factor HIF2alpha and multiple HIF-regulated genes are preferentially expressed in GSCs in comparison to non-stem tumor cells and normal neural progenitors. In tumor specimens, HIF2alpha colocalizes with cancer stem cell markers. Targeting HIFs in GSCs inhibits self-renewal, proliferation, and survival in vitro, and attenuates tumor initiation potential of GSCs in vivo. Analysis of a molecular database reveals that HIF2A expression correlates with poor glioma patient survival. Our results demonstrate that GSCs differentially respond to hypoxia with distinct HIF induction patterns, and HIF2alpha might represent a promising target for antiglioblastoma therapies.
Figures


Similar articles
-
CDH5 is specifically activated in glioblastoma stemlike cells and contributes to vasculogenic mimicry induced by hypoxia.Neuro Oncol. 2013 Jul;15(7):865-79. doi: 10.1093/neuonc/not029. Epub 2013 May 3. Neuro Oncol. 2013. PMID: 23645533 Free PMC article.
-
The HIF-2alpha dependent induction of PAP and adenosine synthesis regulates glioblastoma stem cell function through the A2B adenosine receptor.Int J Biochem Cell Biol. 2014 Apr;49:8-16. doi: 10.1016/j.biocel.2014.01.007. Epub 2014 Jan 13. Int J Biochem Cell Biol. 2014. PMID: 24434023
-
Overexpression of ZNF217 in glioblastoma contributes to the maintenance of glioma stem cells regulated by hypoxia-inducible factors.Lab Invest. 2011 Jul;91(7):1068-78. doi: 10.1038/labinvest.2011.56. Epub 2011 Apr 11. Lab Invest. 2011. PMID: 21483406
-
Hypoxia and hypoxia inducible factors in cancer stem cell maintenance.Curr Top Microbiol Immunol. 2010;345:21-30. doi: 10.1007/82_2010_75. Curr Top Microbiol Immunol. 2010. PMID: 20582533 Review.
-
Glioma stem cell maintenance: the role of the microenvironment.Curr Pharm Des. 2011;17(23):2386-401. doi: 10.2174/138161211797249260. Curr Pharm Des. 2011. PMID: 21827414 Free PMC article. Review.
Cited by
-
Hypoxia-induced HMGB1 promotes glioma stem cells self-renewal and tumorigenicity via RAGE.iScience. 2022 Aug 4;25(9):104872. doi: 10.1016/j.isci.2022.104872. eCollection 2022 Sep 16. iScience. 2022. PMID: 36034219 Free PMC article.
-
The complex interactions between the cellular and non-cellular components of the brain tumor microenvironmental landscape and their therapeutic implications.Front Oncol. 2022 Oct 6;12:1005069. doi: 10.3389/fonc.2022.1005069. eCollection 2022. Front Oncol. 2022. PMID: 36276147 Free PMC article. Review.
-
Drug-resilient Cancer Cell Phenotype Is Acquired via Polyploidization Associated with Early Stress Response Coupled to HIF2α Transcriptional Regulation.Cancer Res Commun. 2024 Mar 7;4(3):691-705. doi: 10.1158/2767-9764.CRC-23-0396. Cancer Res Commun. 2024. PMID: 38385626 Free PMC article.
-
Proteolysis of MOB1 by the ubiquitin ligase praja2 attenuates Hippo signalling and supports glioblastoma growth.Nat Commun. 2013;4:1822. doi: 10.1038/ncomms2791. Nat Commun. 2013. PMID: 23652010 Free PMC article.
-
The Long Non-coding RNA HIF1A-AS2 Facilitates the Maintenance of Mesenchymal Glioblastoma Stem-like Cells in Hypoxic Niches.Cell Rep. 2016 Jun 14;15(11):2500-9. doi: 10.1016/j.celrep.2016.05.018. Epub 2016 Jun 2. Cell Rep. 2016. PMID: 27264189 Free PMC article.
References
-
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006a;444:756–760. - PubMed
-
- Bao S, Wu Q, Sathornsumetee S, Hao Y, Li Z, Hjelmeland AB, Shi Q, McLendon RE, Bigner DD, Rich JN. Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Res. 2006b;66:7843–7848. - PubMed
-
- Beier D, Hau P, Proescholdt M, Lohmeier A, Wischhusen J, Oefner PJ, Aigner L, Brawanski A, Bogdahn U, Beier CP. CD133(+) and CD133(-) glioblastoma-derived cancer stem cells show differential growth characteristics molecular profiles. Cancer Research. 2007;67:4010–4015. - PubMed
-
- Blazek ER, Foutch JL, Maki G. Daoy medulloblastoma cells that express CD133 are radioresistant relative to CD133- cells, and the CD133+ sector is enlarged by hypoxia. Int J Radiat Oncol Biol Phys. 2007;67:1–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

