Visualisation of cyclic and multi-branched molecules with VMD
- PMID: 19473861
- PMCID: PMC3158682
- DOI: 10.1016/j.jmgm.2009.04.010
Visualisation of cyclic and multi-branched molecules with VMD
Abstract
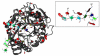
We report the addition of two visualisation algorithms, termed PaperChain and Twister, to the freely available Visual Molecular Dynamics (VMD) package. These algorithms produce visualisations of complex cyclic molecules and multi-branched polysaccharides and are a generalization and optimization of those we previously developed in a standalone package for carbohydrates. PaperChain highlights each ring in a molecular structure with a polygon, which is coloured according to the ring pucker. Twister traces glycosidic bonds with a ribbon that twists according to the relative orientation of successive sugar residues. Combination of these novel algorithms and new ring selection statements with the large set of visualisations already available in VMD allows for unprecedented flexibility in the level of detail displayed for glycoconjugate, glycoprotein and carbohydrate-binding protein structures, as well as other cyclic structures. We highlight the efficacy of these algorithms with selected illustrative examples, clearly demonstrating the value of the new visualisations, not only for structure validation, but also for facilitating insights into molecular structure and mechanism.
Figures
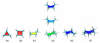
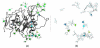



Similar articles
-
Techniques for visualization of carbohydrate molecules.J Mol Graph Model. 2006 Nov;25(3):380-8. doi: 10.1016/j.jmgm.2006.02.007. Epub 2006 Mar 24. J Mol Graph Model. 2006. PMID: 16563824
-
Residue-centric modeling and design of saccharide and glycoconjugate structures.J Comput Chem. 2017 Feb 15;38(5):276-287. doi: 10.1002/jcc.24679. Epub 2016 Nov 30. J Comput Chem. 2017. PMID: 27900782 Free PMC article.
-
Glycan Reader: automated sugar identification and simulation preparation for carbohydrates and glycoproteins.J Comput Chem. 2011 Nov 15;32(14):3135-41. doi: 10.1002/jcc.21886. Epub 2011 Aug 3. J Comput Chem. 2011. PMID: 21815173 Free PMC article.
-
A perspective on the primary and three-dimensional structures of carbohydrates.Carbohydr Res. 2013 Aug 30;378:123-32. doi: 10.1016/j.carres.2013.02.005. Epub 2013 Feb 24. Carbohydr Res. 2013. PMID: 23522728 Review.
-
Molecular dynamics simulations of glycoclusters and glycodendrimers.J Biotechnol. 2002 May;90(3-4):311-37. doi: 10.1016/s1389-0352(01)00072-1. J Biotechnol. 2002. PMID: 12071231 Review.
Cited by
-
Available Instruments for Analyzing Molecular Dynamics Trajectories.Open Biochem J. 2016 Mar 14;10:1-11. doi: 10.2174/1874091X01610010001. eCollection 2016. Open Biochem J. 2016. PMID: 27053964 Free PMC article.
-
Cryptococcus neoformans Capsular GXM Conformation and Epitope Presentation: A Molecular Modelling Study.Molecules. 2020 Jun 7;25(11):2651. doi: 10.3390/molecules25112651. Molecules. 2020. PMID: 32517333 Free PMC article.
-
Conformational comparisons of Pasteurella multocida types B and E and structurally related capsular polysaccharides.Glycobiology. 2023 Oct 29;33(9):745-754. doi: 10.1093/glycob/cwad049. Glycobiology. 2023. PMID: 37334939 Free PMC article.
-
Multiple functions of aromatic-carbohydrate interactions in a processive cellulase examined with molecular simulation.J Biol Chem. 2011 Nov 25;286(47):41028-35. doi: 10.1074/jbc.M111.297713. Epub 2011 Sep 29. J Biol Chem. 2011. PMID: 21965672 Free PMC article.
-
Cross-reactivity of Haemophilus influenzae type a and b polysaccharides: molecular modeling and conjugate immunogenicity studies.Glycoconj J. 2021 Dec;38(6):735-746. doi: 10.1007/s10719-021-10020-0. Epub 2021 Sep 7. Glycoconj J. 2021. PMID: 34491462
References
-
- Richardson JS. The anatomy and taxonomy of protein structure. Advances in Protein Chemistry. 1981;34:167–218. - PubMed
-
- Carson M, Bugg C. Algorithm for ribbon models of proteins. J. Mol. Graph. 1986;4(2):121–122.
-
- Carson M. Ribbon models of macromolecules. J. Mol. Graph. 1987;5(2):103–106.
-
- Crispin M, Stuart DI. Building meaningful models of glycoproteins. Nat. Struct. Mol. Biol. 2007;14:354. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

