In vitro nuclear interactome of the HIV-1 Tat protein
- PMID: 19454010
- PMCID: PMC2702331
- DOI: 10.1186/1742-4690-6-47
In vitro nuclear interactome of the HIV-1 Tat protein
Abstract
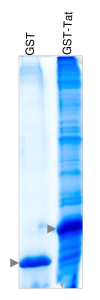
Background: One facet of the complexity underlying the biology of HIV-1 resides not only in its limited number of viral proteins, but in the extensive repertoire of cellular proteins they interact with and their higher-order assembly. HIV-1 encodes the regulatory protein Tat (86-101aa), which is essential for HIV-1 replication and primarily orchestrates HIV-1 provirus transcriptional regulation. Previous studies have demonstrated that Tat function is highly dependent on specific interactions with a range of cellular proteins. However they can only partially account for the intricate molecular mechanisms underlying the dynamics of proviral gene expression. To obtain a comprehensive nuclear interaction map of Tat in T-cells, we have designed a proteomic strategy based on affinity chromatography coupled with mass spectrometry.
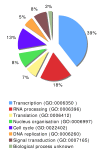
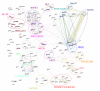
Results: Our approach resulted in the identification of a total of 183 candidates as Tat nuclear partners, 90% of which have not been previously characterised. Subsequently we applied in silico analysis, to validate and characterise our dataset which revealed that the Tat nuclear interactome exhibits unique signature(s). First, motif composition analysis highlighted that our dataset is enriched for domains mediating protein, RNA and DNA interactions, and helicase and ATPase activities. Secondly, functional classification and network reconstruction clearly depicted Tat as a polyvalent protein adaptor and positioned Tat at the nexus of a densely interconnected interaction network involved in a range of biological processes which included gene expression regulation, RNA biogenesis, chromatin structure, chromosome organisation, DNA replication and nuclear architecture.
Conclusion: We have completed the in vitro Tat nuclear interactome and have highlighted its modular network properties and particularly those involved in the coordination of gene expression by Tat. Ultimately, the highly specialised set of molecular interactions identified will provide a framework to further advance our understanding of the mechanisms of HIV-1 proviral gene silencing and activation.
Figures

Similar articles
-
Nucleolar protein trafficking in response to HIV-1 Tat: rewiring the nucleolus.PLoS One. 2012;7(11):e48702. doi: 10.1371/journal.pone.0048702. Epub 2012 Nov 15. PLoS One. 2012. PMID: 23166591 Free PMC article.
-
HIV-1 Tat phosphorylation on Ser-16 residue modulates HIV-1 transcription.Retrovirology. 2018 May 23;15(1):39. doi: 10.1186/s12977-018-0422-5. Retrovirology. 2018. PMID: 29792216 Free PMC article.
-
HIV latency reversing agents act through Tat post translational modifications.Retrovirology. 2018 May 11;15(1):36. doi: 10.1186/s12977-018-0421-6. Retrovirology. 2018. PMID: 29751762 Free PMC article.
-
Tat is a multifunctional viral protein that modulates cellular gene expression and functions.Oncotarget. 2017 Apr 18;8(16):27569-27581. doi: 10.18632/oncotarget.15174. Oncotarget. 2017. PMID: 28187438 Free PMC article. Review.
-
The Tat/P-TEFb Protein-Protein Interaction Determining Transcriptional Activation of HIV.Curr Pharm Des. 2017;23(28):4091-4097. doi: 10.2174/1381612823666170710164148. Curr Pharm Des. 2017. PMID: 28699519 Review.
Cited by
-
The Road Less Traveled: HIV's Use of Alternative Routes through Cellular Pathways.J Virol. 2015 May;89(10):5204-12. doi: 10.1128/JVI.03684-14. Epub 2015 Mar 11. J Virol. 2015. PMID: 25762730 Free PMC article. Review.
-
Macrophage signaling in HIV-1 infection.Retrovirology. 2010 Apr 9;7:34. doi: 10.1186/1742-4690-7-34. Retrovirology. 2010. PMID: 20380698 Free PMC article. Review.
-
HIV-1 Tat protein induces DNA damage in human peripheral blood B-lymphocytes via mitochondrial ROS production.Redox Biol. 2018 May;15:97-108. doi: 10.1016/j.redox.2017.11.024. Epub 2017 Dec 7. Redox Biol. 2018. PMID: 29220699 Free PMC article.
-
Unbiased proteomic analysis of proteins interacting with the HIV-1 5'LTR sequence: role of the transcription factor Meis.Nucleic Acids Res. 2012 Nov;40(21):e168. doi: 10.1093/nar/gks733. Epub 2012 Aug 16. Nucleic Acids Res. 2012. PMID: 22904091 Free PMC article.
-
Regulation of Tat acetylation and transactivation activity by the microtubule-associated deacetylase HDAC6.J Biol Chem. 2011 Mar 18;286(11):9280-6. doi: 10.1074/jbc.M110.208884. Epub 2011 Jan 10. J Biol Chem. 2011. PMID: 21220424 Free PMC article.
References
-
- Bannwarth S, Gatignol A. HIV-1 TAR RNA: the target of molecular interactions between the virus and its host. Curr HIV Res. 2005;3:61–71. - PubMed
-
- Nekhai S, Jeang KT. Transcriptional and post-transcriptional regulation of HIV-1 gene expression: role of cellular factors for Tat and Rev. Future Microbiol. 2006;1:417–426. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

