Thermostabilization of the neurotensin receptor NTS1
- PMID: 19422831
- PMCID: PMC2696590
- DOI: 10.1016/j.jmb.2009.04.068
Thermostabilization of the neurotensin receptor NTS1
Abstract
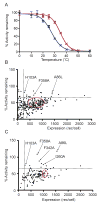
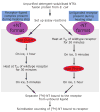
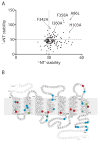
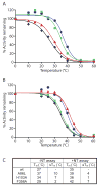
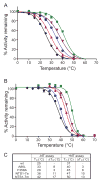
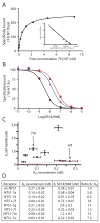
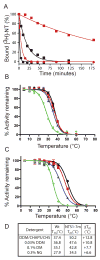
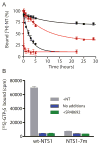
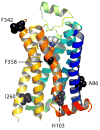
Structural studies on G-protein-coupled receptors have been hampered for many years by their instability in detergent solution and by the number of potential conformations that receptors can adopt. Recently, the structures of the beta(1) and beta(2) adrenergic receptors and the adenosine A(2a) receptor were determined in the antagonist-bound state, a receptor conformation that is thought to be more stable than the agonist-bound state. In contrast to these receptors, the neurotensin (NT) receptor NTS1 is much less stable in detergent solution. We have therefore used a systematic mutational approach coupled with activity assays to identify receptor mutants suitable for crystallization, both alone and in complex with the peptide agonist NT. The best receptor mutant NTS1-7m contained four point mutations. It showed increased stability compared to the wild-type receptor, in the absence of ligand, after solubilization with a variety of detergents. In addition, NTS1-7m bound to NT was more stable than unliganded NTS1-7m. Of the four thermostabilizing mutations, only one residue (A86L) is predicted to be in the lipid environment. In contrast, I260A appears to be buried within the transmembrane helix bundle, F342A may form a distant part of the putative ligand-binding site, whereas F358A is likely to be in a region that is important for receptor activation. NTS1-7m binds NT with a similar affinity for the wild-type receptor. However, agonist dissociation was slower, and NTS1-7m activated G-proteins poorly. The affinity of NTS1-7m for the antagonist SR48692 was also lower than that of the wild-type receptor. Thus, we have successfully stabilized NTS1 in an agonist-binding conformation that does not efficiently couple to G-proteins.
Figures
Similar articles
-
Optimization and 13CH3 methionine labeling of a signaling competent neurotensin receptor 1 variant for NMR studies.Biochim Biophys Acta Biomembr. 2018 Jun;1860(6):1372-1383. doi: 10.1016/j.bbamem.2018.03.020. Epub 2018 Mar 26. Biochim Biophys Acta Biomembr. 2018. PMID: 29596791
-
Stability of the neurotensin receptor NTS1 free in detergent solution and immobilized to affinity resin.PLoS One. 2010 Sep 7;5(9):e12579. doi: 10.1371/journal.pone.0012579. PLoS One. 2010. PMID: 20830205 Free PMC article.
-
Impaired G protein coupling of the neurotensin receptor 1 by mutations in extracellular loop 3.Eur J Pharmacol. 2001 Dec 14;433(1):63-71. doi: 10.1016/s0014-2999(01)01496-0. Eur J Pharmacol. 2001. PMID: 11755135
-
Molecular and cellular regulation of neurotensin receptor under acute and chronic agonist stimulation.Peptides. 2006 Oct;27(10):2493-501. doi: 10.1016/j.peptides.2006.04.029. Epub 2006 Aug 4. Peptides. 2006. PMID: 16889873 Review.
-
Functional domains of the subtype 1 neurotensin receptor (NTS1).Peptides. 2006 Oct;27(10):2461-8. doi: 10.1016/j.peptides.2006.02.013. Epub 2006 Aug 9. Peptides. 2006. PMID: 16901586 Review.
Cited by
-
Characterisation of a cell-free synthesised G-protein coupled receptor.Sci Rep. 2017 Apr 24;7(1):1094. doi: 10.1038/s41598-017-01227-z. Sci Rep. 2017. PMID: 28439124 Free PMC article.
-
Micro-scale and rapid expression screening of highly expressed and/or stable membrane protein variants in Saccharomyces cerevisiae.Protein Sci. 2016 Oct;25(10):1863-72. doi: 10.1002/pro.2993. Epub 2016 Aug 13. Protein Sci. 2016. PMID: 27479358 Free PMC article.
-
Extra-helical binding site of a glucagon receptor antagonist.Nature. 2016 May 12;533(7602):274-7. doi: 10.1038/nature17414. Epub 2016 Apr 25. Nature. 2016. PMID: 27111510
-
Stabilization of functional recombinant cannabinoid receptor CB(2) in detergent micelles and lipid bilayers.PLoS One. 2012;7(10):e46290. doi: 10.1371/journal.pone.0046290. Epub 2012 Oct 3. PLoS One. 2012. PMID: 23056277 Free PMC article.
-
A class of mild surfactants that keep integral membrane proteins water-soluble for functional studies and crystallization.Mol Membr Biol. 2011 Apr;28(3):171-81. doi: 10.3109/09687688.2011.552440. Epub 2011 Feb 14. Mol Membr Biol. 2011. PMID: 21314479 Free PMC article.
References
-
- Warne T, Serrano-Vega MJ, Tate CG, Schertler GF. Development and crystallization of a minimal thermostabilized G protein-coupled receptor. Protein Exp Purif. 2009;65:204–213. - PubMed
-
- Deisenhofer J, Epp O, Miki K, Huber R, Michel H. Structure of the protein subunits in the photosynthetic reaction centre of Rhodopseudomonas viridis at 3Å resolution. Nature. 1985;318:618–624. - PubMed
-
- Weiss MS, Schulz GE. Structure of porin refined at 1.8 Å resolution. J Mol Biol. 1992;227:493–509. - PubMed
-
- Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Å. Science. 1996;272:1136–44. - PubMed
-
- Xia D, Yu CA, Kim H, Xia JZ, Kachurin AM, Zhang L, Yu L, Deisenhofer J. Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science. 1997;277:60–66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

