The nucleoporins Nup170p and Nup157p are essential for nuclear pore complex assembly
- PMID: 19414608
- PMCID: PMC2700392
- DOI: 10.1083/jcb.200810029
The nucleoporins Nup170p and Nup157p are essential for nuclear pore complex assembly
Abstract
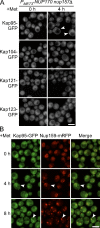
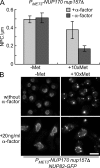
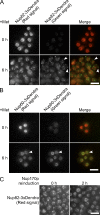
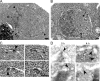
We have established that two homologous nucleoporins, Nup170p and Nup157p, play an essential role in the formation of nuclear pore complexes (NPCs) in Saccharomyces cerevisiae. By regulating their synthesis, we showed that the loss of these nucleoporins triggers a decrease in NPCs caused by a halt in new NPC assembly. Preexisting NPCs are ultimately lost by dilution as cells grow, causing the inhibition of nuclear transport and the loss of viability. Significantly, the loss of Nup170p/Nup157p had distinct effects on the assembly of different architectural components of the NPC. Nucleoporins (nups) positioned on the cytoplasmic face of the NPC rapidly accumulated in cytoplasmic foci. These nup complexes could be recruited into new NPCs after reinitiation of Nup170p synthesis, and may represent a physiological intermediate. Loss of Nup170p/Nup157p also caused core and nucleoplasmically positioned nups to accumulate in NPC-like structures adjacent to the inner nuclear membrane, which suggests that these nucleoporins are required for formation of the pore membrane and the incorporation of cytoplasmic nups into forming NPCs.
Figures

Comment in
-
Piecing together nuclear pore complex assembly during interphase.J Cell Biol. 2009 May 4;185(3):377-9. doi: 10.1083/jcb.200904022. J Cell Biol. 2009. PMID: 19414605 Free PMC article.
Similar articles
-
Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p.J Cell Biol. 1995 Dec;131(5):1133-48. doi: 10.1083/jcb.131.5.1133. J Cell Biol. 1995. PMID: 8522578 Free PMC article.
-
The integral membrane protein Pom34p functionally links nucleoporin subcomplexes.Genetics. 2006 Mar;172(3):1441-57. doi: 10.1534/genetics.105.052068. Epub 2005 Dec 15. Genetics. 2006. PMID: 16361228 Free PMC article.
-
Assembly of Nsp1 nucleoporins provides insight into nuclear pore complex gating.PLoS Comput Biol. 2014 Mar 13;10(3):e1003488. doi: 10.1371/journal.pcbi.1003488. eCollection 2014 Mar. PLoS Comput Biol. 2014. PMID: 24626154 Free PMC article.
-
[Nuclear pores: from yeast to higher eukaryotes].J Soc Biol. 2002;196(4):349-54. J Soc Biol. 2002. PMID: 12645306 Review. French.
-
Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions.Nat Rev Mol Cell Biol. 2012 Nov;13(11):687-99. doi: 10.1038/nrm3461. Nat Rev Mol Cell Biol. 2012. PMID: 23090414 Review.
Cited by
-
The SUN protein Mps3 is required for spindle pole body insertion into the nuclear membrane and nuclear envelope homeostasis.PLoS Genet. 2011 Nov;7(11):e1002365. doi: 10.1371/journal.pgen.1002365. Epub 2011 Nov 17. PLoS Genet. 2011. PMID: 22125491 Free PMC article.
-
The chaperone DNAJB6 surveils FG-nucleoporins and is required for interphase nuclear pore complex biogenesis.Nat Cell Biol. 2022 Nov;24(11):1584-1594. doi: 10.1038/s41556-022-01010-x. Epub 2022 Oct 27. Nat Cell Biol. 2022. PMID: 36302971
-
Uip4p modulates nuclear pore complex function in Saccharomyces cerevisiae.Nucleus. 2022 Dec;13(1):79-93. doi: 10.1080/19491034.2022.2034286. Nucleus. 2022. PMID: 35171083 Free PMC article.
-
Trafficking and/or division: Distinct roles of nucleoporins based on their location within the nuclear pore complex.RNA Biol. 2022;19(1):650-661. doi: 10.1080/15476286.2022.2067711. Epub 2021 Dec 31. RNA Biol. 2022. PMID: 35491934 Free PMC article.
-
Dunking into the Lipid Bilayer: How Direct Membrane Binding of Nucleoporins Can Contribute to Nuclear Pore Complex Structure and Assembly.Cells. 2021 Dec 20;10(12):3601. doi: 10.3390/cells10123601. Cells. 2021. PMID: 34944108 Free PMC article. Review.
References
-
- Aitchison J.D., Rout M.P., Marelli M., Blobel G., Wozniak R.W. 1995. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p.J. Cell Biol. 131:1133–1148 - PMC - PubMed
-
- Alber F., Dokudovskaya S., Veenhoff L.M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B.T., et al. 2007a. Determining the architectures of macromolecular assemblies.Nature. 450:683–694 - PubMed
-
- Alber F., Dokudovskaya S., Veenhoff L.M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B.T., et al. 2007b. The molecular architecture of the nuclear pore complex.Nature. 450:695–701 - PubMed
-
- Amberg D.C., Goldstein A.L., Cole C.N. 1992. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA.Genes Dev. 6:1173–1189 - PubMed
-
- Antonin W., Franz C., Haselmann U., Antony C., Mattaj I.W. 2005. The integral membrane nucleoporin pom121 functionally links nuclear pore complex assembly and nuclear envelope formation.Mol. Cell. 17:83–92 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

