Sir2 paralogues cooperate to regulate virulence genes and antigenic variation in Plasmodium falciparum
- PMID: 19402747
- PMCID: PMC2672602
- DOI: 10.1371/journal.pbio.1000084
Sir2 paralogues cooperate to regulate virulence genes and antigenic variation in Plasmodium falciparum
Abstract
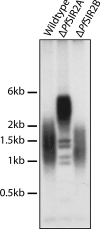
Cytoadherance of Plasmodium falciparum-infected erythrocytes in the brain, organs and peripheral microvasculature is linked to morbidity and mortality associated with severe malaria. Parasite-derived P. falciparum Erythrocyte Membrane Protein 1 (PfEMP1) molecules displayed on the erythrocyte surface are responsible for cytoadherance and undergo antigenic variation in the course of an infection. Antigenic variation of PfEMP1 is achieved by in situ switching and mutually exclusive transcription of the var gene family, a process that is controlled by epigenetic mechanisms. Here we report characterisation of the P. falciparum silent information regulator's A and B (PfSir2A and PfSir2B) and their involvement in mutual exclusion and silencing of the var gene repertoire. Analysis of P. falciparum parasites lacking either PfSir2A or PfSir2B shows that these NAD(+)-dependent histone deacetylases are required for silencing of different var gene subsets classified by their conserved promoter type. We also demonstrate that in the absence of either of these molecules mutually exclusive expression of var genes breaks down. We show that var gene silencing originates within the promoter and PfSir2 paralogues are involved in cis spreading of silenced chromatin into adjacent regions. Furthermore, parasites lacking PfSir2A but not PfSir2B have considerably longer telomeric repeats, demonstrating a role for this molecule in telomeric end protection. This work highlights the pivotal but distinct role for both PfSir2 paralogues in epigenetic silencing of P. falciparum virulence genes and the control of pathogenicity of malaria infection.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures

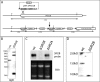

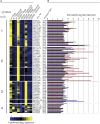
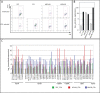

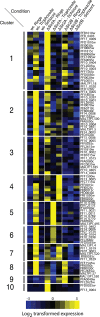
Similar articles
-
The Plasmodium falciparum histone methyltransferase PfSET10 is dispensable for the regulation of antigenic variation and gene expression in blood-stage parasites.mSphere. 2024 Nov 21;9(11):e0054624. doi: 10.1128/msphere.00546-24. Epub 2024 Oct 24. mSphere. 2024. PMID: 39445826 Free PMC article.
-
A var gene promoter controls allelic exclusion of virulence genes in Plasmodium falciparum malaria.Nature. 2006 Feb 23;439(7079):1004-8. doi: 10.1038/nature04407. Epub 2005 Dec 28. Nature. 2006. PMID: 16382237
-
Febrile temperature causes transcriptional downregulation of Plasmodium falciparum Sirtuins through Hsp90-dependent epigenetic modification.Mol Microbiol. 2021 May;115(5):1025-1038. doi: 10.1111/mmi.14692. Epub 2021 Feb 13. Mol Microbiol. 2021. PMID: 33538363
-
Antigenic variation in Plasmodium falciparum.Annu Rev Microbiol. 2008;62:445-70. doi: 10.1146/annurev.micro.61.080706.093134. Annu Rev Microbiol. 2008. PMID: 18785843 Review.
-
[Var gene family and antigen variation in Plasmodium falciparum].Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi. 2010 Apr;28(2):153-6. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi. 2010. PMID: 20666324 Review. Chinese.
Cited by
-
Dynamic Chromatin Structure and Epigenetics Control the Fate of Malaria Parasites.Trends Genet. 2021 Jan;37(1):73-85. doi: 10.1016/j.tig.2020.09.003. Epub 2020 Sep 25. Trends Genet. 2021. PMID: 32988634 Free PMC article. Review.
-
Transcription of the var genes from a freshly-obtained field isolate of Plasmodium falciparum shows more variable switching patterns than long laboratory-adapted isolates.Malar J. 2015 Feb 7;14:66. doi: 10.1186/s12936-015-0565-y. Malar J. 2015. PMID: 25889871 Free PMC article.
-
Solute restriction reveals an essential role for clag3-associated channels in malaria parasite nutrient acquisition.Mol Pharmacol. 2012 Dec;82(6):1104-14. doi: 10.1124/mol.112.081224. Epub 2012 Sep 4. Mol Pharmacol. 2012. PMID: 22949525 Free PMC article.
-
In silico identification of inhibitors against Plasmodium falciparum histone deacetylase 1 (PfHDAC-1).J Mol Model. 2018 Aug 14;24(9):232. doi: 10.1007/s00894-018-3761-1. J Mol Model. 2018. PMID: 30109440
-
Chromatin-mediated epigenetic regulation in the malaria parasite Plasmodium falciparum.Eukaryot Cell. 2010 Aug;9(8):1138-49. doi: 10.1128/EC.00036-10. Epub 2010 May 7. Eukaryot Cell. 2010. PMID: 20453074 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

