The role of disordered ribosomal protein extensions in the early steps of eubacterial 50 S ribosomal subunit assembly
- PMID: 19399222
- PMCID: PMC2672003
- DOI: 10.3390/ijms10030817
The role of disordered ribosomal protein extensions in the early steps of eubacterial 50 S ribosomal subunit assembly
Abstract
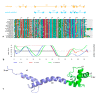
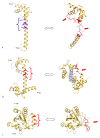
Although during the past decade research has shown the functional importance of disorder in proteins, many of the structural and dynamics properties of intrinsically unstructured proteins (IUPs) remain to be elucidated. This review is focused on the role of the extensions of the ribosomal proteins in the early steps of the assembly of the eubacterial 50 S subunit. The recent crystallographic structures of the ribosomal particles have revealed the picture of a complex assembly pathway that condenses the rRNA and the ribosomal proteins into active ribosomes. However, little is know about the molecular mechanisms of this process. It is thought that the long basic r-protein extensions that penetrate deeply into the subunit cores play a key role through disorder-order transitions and/or co-folding mechanisms. A current view is that such structural transitions may facilitate the proper rRNA folding. In this paper, the structures of the proteins L3, L4, L13, L20, L22 and L24 that have been experimentally found to be essential for the first steps of ribosome assembly have been compared. On the basis of their structural and dynamics properties, three categories of extensions have been identified. Each of them seems to play a distinct function. Among them, only the coil-helix transition that occurs in a phylogenetically conserved cluster of basic residues of the L20 extension appears to be strictly required for the large subunit assembly in eubacteria. The role of alpha helix-coil transitions in 23 S RNA folding is discussed in the light of the calcium binding protein calmodulin that shares many structural and dynamics properties with L20.
Keywords: Flexibility; calmodulin; electrostatic; helix unwinding; helix-coil; linker; structural transitions; unfolding.
Figures
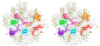

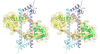



Similar articles
-
Visualization of protein-nucleic acid interactions involved in the in vitro assembly of the Escherichia coli 50 S ribosomal subunit.J Mol Biol. 1994 Jan 28;235(4):1239-50. doi: 10.1006/jmbi.1994.1077. J Mol Biol. 1994. PMID: 8308887
-
Coexistence of two protein folding states in the crystal structure of ribosomal protein L20.EMBO Rep. 2006 Oct;7(10):1013-8. doi: 10.1038/sj.embor.7400803. Epub 2006 Sep 15. EMBO Rep. 2006. PMID: 16977336 Free PMC article.
-
The N-terminal extension of yeast ribosomal protein L8 is involved in two major remodeling events during late nuclear stages of 60S ribosomal subunit assembly.RNA. 2016 Sep;22(9):1386-99. doi: 10.1261/rna.055798.115. Epub 2016 Jul 7. RNA. 2016. PMID: 27390266 Free PMC article.
-
Critical steps in the assembly process of the bacterial 50S ribosomal subunit.Nucleic Acids Res. 2024 May 8;52(8):4111-4123. doi: 10.1093/nar/gkae199. Nucleic Acids Res. 2024. PMID: 38554105 Free PMC article. Review.
-
Eukaryotic ribosomal proteins lacking a eubacterial counterpart: important players in ribosomal function.Mol Microbiol. 2006 Mar;59(6):1651-63. doi: 10.1111/j.1365-2958.2006.05054.x. Mol Microbiol. 2006. PMID: 16553873 Review.
Cited by
-
The mitochondrial ribosomal protein L13 is critical for the structural and functional integrity of the mitochondrion in Plasmodium falciparum.J Biol Chem. 2018 May 25;293(21):8128-8137. doi: 10.1074/jbc.RA118.002552. Epub 2018 Apr 6. J Biol Chem. 2018. PMID: 29626096 Free PMC article.
-
Spatially Enriched Paralog Rearrangements Argue Functionally Diverse Ribosomes Arise during Cold Acclimation in Arabidopsis.Int J Mol Sci. 2021 Jun 7;22(11):6160. doi: 10.3390/ijms22116160. Int J Mol Sci. 2021. PMID: 34200446 Free PMC article.
-
Neuron-Like Networks Between Ribosomal Proteins Within the Ribosome.Sci Rep. 2016 May 26;6:26485. doi: 10.1038/srep26485. Sci Rep. 2016. PMID: 27225526 Free PMC article.
-
Moonlighting Proteins in the Fuzzy Logic of Cellular Metabolism.Molecules. 2020 Jul 29;25(15):3440. doi: 10.3390/molecules25153440. Molecules. 2020. PMID: 32751110 Free PMC article. Review.
-
Dissecting macromolecular recognition sites in ribosome: implication to its self-assembly.RNA Biol. 2019 Sep;16(9):1300-1312. doi: 10.1080/15476286.2019.1629767. Epub 2019 Jun 17. RNA Biol. 2019. PMID: 31179876 Free PMC article.
References
-
- Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999;293:321–331. - PubMed
-
- Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner E, Obradovic Z. Intrinsically disordered protein. J. Mol. Graph. Model. 2001;19:26–59. - PubMed
-
- Tompa P. Intrinsically unstructured proteins. Trends Biochem. Sci. 2002;27:527–533. - PubMed
-
- Fink A. Natively unfolded proteins. Curr. Opin. Struct. Biol. 2005;15:35–41. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

