Molecular dynamics flexible fitting: a practical guide to combine cryo-electron microscopy and X-ray crystallography
- PMID: 19398010
- PMCID: PMC2753685
- DOI: 10.1016/j.ymeth.2009.04.005
Molecular dynamics flexible fitting: a practical guide to combine cryo-electron microscopy and X-ray crystallography
Abstract
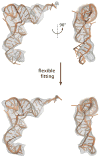
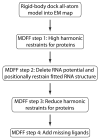
Hybrid computational methods for combining structural data from different sources and resolutions are becoming an essential part of structural biology, especially as the field moves toward the study of large macromolecular assemblies. We have developed the molecular dynamics flexible fitting (MDFF) method for combining high-resolution atomic structures with cryo-electron microscopy (cryo-EM) maps, that results in atomic models representing the conformational state captured by cryo-EM. The method has been applied successfully to the ribosome, a ribonucleoprotein complex responsible for protein synthesis. MDFF involves a molecular dynamics simulation in which a guiding potential, based on the cryo-EM map, is added to the standard force field. Forces proportional to the gradient of the density map guide an atomic structure, available from X-ray crystallography, into high-density regions of a cryo-EM map. In this paper we describe the necessary steps to set up, run, and analyze MDFF simulations and the software packages that implement the corresponding functionalities.
Figures
Similar articles
-
Cryo-electron microscopy modeling by the molecular dynamics flexible fitting method.Biopolymers. 2012 Sep;97(9):678-86. doi: 10.1002/bip.22042. Biopolymers. 2012. PMID: 22696404 Free PMC article.
-
Applications of the molecular dynamics flexible fitting method.J Struct Biol. 2011 Mar;173(3):420-7. doi: 10.1016/j.jsb.2010.09.024. Epub 2010 Oct 12. J Struct Biol. 2011. PMID: 20932910 Free PMC article.
-
Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics.Structure. 2008 May;16(5):673-83. doi: 10.1016/j.str.2008.03.005. Structure. 2008. PMID: 18462672 Free PMC article.
-
Molecular simulations of the ribosome and associated translation factors.Curr Opin Struct Biol. 2018 Apr;49:27-35. doi: 10.1016/j.sbi.2017.11.003. Epub 2017 Dec 1. Curr Opin Struct Biol. 2018. PMID: 29202442 Review.
-
Ribosomes and cryo-EM: a duet.Curr Opin Struct Biol. 2018 Oct;52:1-7. doi: 10.1016/j.sbi.2018.07.001. Epub 2018 Jul 14. Curr Opin Struct Biol. 2018. PMID: 30015201 Review.
Cited by
-
Structure of a group II intron in complex with its reverse transcriptase.Nat Struct Mol Biol. 2016 Jun;23(6):549-57. doi: 10.1038/nsmb.3220. Epub 2016 May 2. Nat Struct Mol Biol. 2016. PMID: 27136327 Free PMC article.
-
Structure of the human 26S proteasome at a resolution of 3.9 Å.Proc Natl Acad Sci U S A. 2016 Jul 12;113(28):7816-21. doi: 10.1073/pnas.1608050113. Epub 2016 Jun 24. Proc Natl Acad Sci U S A. 2016. PMID: 27342858 Free PMC article.
-
Subunit folds and maturation pathway of a dsRNA virus capsid.Structure. 2013 Aug 6;21(8):1374-83. doi: 10.1016/j.str.2013.06.007. Epub 2013 Jul 25. Structure. 2013. PMID: 23891288 Free PMC article.
-
The bacteriophage ϕ29 tail possesses a pore-forming loop for cell membrane penetration.Nature. 2016 Jun 23;534(7608):544-7. doi: 10.1038/nature18017. Epub 2016 Jun 15. Nature. 2016. PMID: 27309813
-
Cryo-electron microscopy modeling by the molecular dynamics flexible fitting method.Biopolymers. 2012 Sep;97(9):678-86. doi: 10.1002/bip.22042. Biopolymers. 2012. PMID: 22696404 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

