MicroRNA-320 is involved in the regulation of cardiac ischemia/reperfusion injury by targeting heat-shock protein 20
- PMID: 19380620
- PMCID: PMC2746735
- DOI: 10.1161/CIRCULATIONAHA.108.814145
MicroRNA-320 is involved in the regulation of cardiac ischemia/reperfusion injury by targeting heat-shock protein 20
Abstract
Background: Recent studies have identified critical roles for microRNAs (miRNAs) in a variety of cellular processes, including regulation of cardiomyocyte death. However, the signature of miRNA expression and possible roles of miRNA in the ischemic heart have been less well studied.
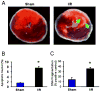
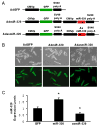
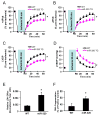
Methods and results: We performed miRNA arrays to detect the expression pattern of miRNAs in murine hearts subjected to ischemia/reperfusion (I/R) in vivo and ex vivo. Surprisingly, we found that only miR-320 expression was significantly decreased in the hearts on I/R in vivo and ex vivo. This was further confirmed by TaqMan real-time polymerase chain reaction. Gain-of-function and loss-of-function approaches were employed in cultured adult rat cardiomyocytes to investigate the functional roles of miR-320. Overexpression of miR-320 enhanced cardiomyocyte death and apoptosis, whereas knockdown was cytoprotective, on simulated I/R. Furthermore, transgenic mice with cardiac-specific overexpression of miR-320 revealed an increased extent of apoptosis and infarction size in the hearts on I/R in vivo and ex vivo relative to the wild-type controls. Conversely, in vivo treatment with antagomir-320 reduced infarction size relative to the administration of mutant antagomir-320 and saline controls. Using TargetScan software and proteomic analysis, we identified heat-shock protein 20 (Hsp20), a known cardioprotective protein, as an important candidate target for miR-320. This was validated experimentally by utilizing a luciferase/GFP reporter activity assay and examining the expression of Hsp20 on miR-320 overexpression and knockdown in cardiomyocytes.
Conclusions: Our data demonstrate that miR-320 is involved in the regulation of I/R-induced cardiac injury and dysfunction via antithetical regulation of Hsp20. Thus, miR-320 may constitute a new therapeutic target for ischemic heart diseases.
Conflict of interest statement
Figures









Similar articles
-
Novel cardioprotective role of a small heat-shock protein, Hsp20, against ischemia/reperfusion injury.Circulation. 2005 Apr 12;111(14):1792-9. doi: 10.1161/01.CIR.0000160851.41872.C6. Epub 2005 Apr 4. Circulation. 2005. PMID: 15809372
-
MicroRNA-494 targeting both proapoptotic and antiapoptotic proteins protects against ischemia/reperfusion-induced cardiac injury.Circulation. 2010 Sep 28;122(13):1308-18. doi: 10.1161/CIRCULATIONAHA.110.964684. Epub 2010 Sep 13. Circulation. 2010. PMID: 20837890 Free PMC article.
-
Gene transfer of heat-shock protein 20 protects against ischemia/reperfusion injury in rat hearts.Acta Pharmacol Sin. 2005 Oct;26(10):1193-200. doi: 10.1111/j.1745-7254.2005.00139.x. Acta Pharmacol Sin. 2005. PMID: 16174435
-
The role of microRNA in modulating myocardial ischemia-reperfusion injury.Physiol Genomics. 2011 May 1;43(10):534-42. doi: 10.1152/physiolgenomics.00130.2010. Epub 2010 Oct 19. Physiol Genomics. 2011. PMID: 20959496 Review.
-
PKA phosphorylation of the small heat-shock protein Hsp20 enhances its cardioprotective effects.Biochem Soc Trans. 2012 Feb;40(1):210-4. doi: 10.1042/BST20110673. Biochem Soc Trans. 2012. PMID: 22260692 Free PMC article. Review.
Cited by
-
Deficiency of a novel lncRNA-HRAT protects against myocardial ischemia reperfusion injury by targeting miR-370-3p/RNF41 pathway.Front Cardiovasc Med. 2022 Sep 12;9:951463. doi: 10.3389/fcvm.2022.951463. eCollection 2022. Front Cardiovasc Med. 2022. PMID: 36172578 Free PMC article.
-
MicroRNAs in Acute ST Elevation Myocardial Infarction-A New Tool for Diagnosis and Prognosis: Therapeutic Implications.Int J Mol Sci. 2021 Apr 30;22(9):4799. doi: 10.3390/ijms22094799. Int J Mol Sci. 2021. PMID: 33946541 Free PMC article. Review.
-
MicroRNAs in heart development.Curr Top Dev Biol. 2012;100:279-317. doi: 10.1016/B978-0-12-387786-4.00009-9. Curr Top Dev Biol. 2012. PMID: 22449848 Free PMC article. Review.
-
MicroRNA-93 inhibits ischemia-reperfusion induced cardiomyocyte apoptosis by targeting PTEN.Oncotarget. 2016 May 17;7(20):28796-805. doi: 10.18632/oncotarget.8941. Oncotarget. 2016. PMID: 27119510 Free PMC article.
-
Research hotspots and development trends of microRNA in ischemia-reperfusion: network analysis of academic journals oriented by bibliometric and visualization.Ann Transl Med. 2022 Dec;10(24):1321. doi: 10.21037/atm-22-5677. Ann Transl Med. 2022. PMID: 36660677 Free PMC article.
References
-
- Cannon RO., 3rd Mechanisms, management and future directions for reperfusion injury after acute myocardial infarction. Nat Clin Pract Cardiovasc Med. 2005;2:88–94. - PubMed
-
- Abbate A, Bussani R, Amin MS, Vetrovec GW, Baldi A. Acute myocardial infarction and heart failure: role of apoptosis. Int J Biochem Cell Biol. 2006;38:1834–1840. - PubMed
-
- Cokkinos DV, Pantos C. Myocardial protection in man--from research concept to clinical practice. Heart Fail Rev. 2007;12:345–362. - PubMed
-
- Latronico MV, Catalucci D, Condorelli G. Emerging role of microRNAs in cardiovascular biology. Circ Res. 2007;101:1225–1236. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

