The Human Scavenger Receptor CD36: glycosylation status and its role in trafficking and function
- PMID: 19369259
- PMCID: PMC2713513
- DOI: 10.1074/jbc.M109.007849
The Human Scavenger Receptor CD36: glycosylation status and its role in trafficking and function
Abstract
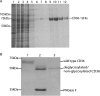
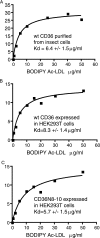
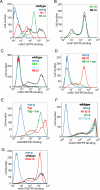
Human CD36 is a class B scavenger receptor expressed in a variety of cell types such as macrophage and adipocytes. This plasma membrane glycoprotein has a wide range of ligands including oxidized low density lipoprotein and long chain fatty acids which involves the receptor in diseases such as atherosclerosis and insulin resistance. CD36 is heavily modified post-translationally by N-linked glycosylation, and 10 putative glycosylation sites situated in the large extracellular loop of the protein have been identified; however, their utilization and role in the folding and function of the protein have not been characterized. Using mass spectrometry on purified and peptide N-glycosidase F-deglycosylated CD36 and also by comparing the electrophoretic mobility of different glycosylation site mutants, we have determined that 9 of the 10 sites can be modified by glycosylation. Flow cytometric analysis of the different glycosylation mutants expressed in mammalian cells established that glycosylation is necessary for trafficking to the plasma membrane. Minimally glycosylated mutants that supported trafficking were identified and indicated the importance of carboxyl-terminal sites Asn-247, Asn-321, and Asn-417. However, unlike SRBI, no individual site was found to be essential for proper trafficking of CD36. Surprisingly, these minimally glycosylated mutants appear to be predominantly core-glycosylated, indicating that mature glycosylation is not necessary for surface expression in mammalian cells. The data also show that neither the nature nor the pattern of glycosylation is relevant to binding of modified low density lipoprotein.
Figures


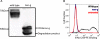

Similar articles
-
The efficient cellular uptake of high density lipoprotein lipids via scavenger receptor class B type I requires not only receptor-mediated surface binding but also receptor-specific lipid transfer mediated by its extracellular domain.J Biol Chem. 1998 Oct 9;273(41):26338-48. doi: 10.1074/jbc.273.41.26338. J Biol Chem. 1998. PMID: 9756864
-
The modulation of the hexosamine biosynthetic pathway impacts the localization of CD36 in macrophages.Acta Biochim Pol. 2024 Jul 8;71:13004. doi: 10.3389/abp.2024.13004. eCollection 2024. Acta Biochim Pol. 2024. PMID: 39041003 Free PMC article.
-
Human CD36 is a high affinity receptor for the native lipoproteins HDL, LDL, and VLDL.J Lipid Res. 1998 Apr;39(4):777-88. J Lipid Res. 1998. PMID: 9555943
-
The enigmatic membrane fatty acid transporter CD36: New insights into fatty acid binding and their effects on uptake of oxidized LDL.Prostaglandins Leukot Essent Fatty Acids. 2018 Nov;138:64-70. doi: 10.1016/j.plefa.2016.05.005. Epub 2016 May 20. Prostaglandins Leukot Essent Fatty Acids. 2018. PMID: 27288302 Review.
-
CD36 in Atherosclerosis: Pathophysiological Mechanisms and Therapeutic Implications.Curr Atheroscler Rep. 2020 Aug 9;22(10):59. doi: 10.1007/s11883-020-00870-8. Curr Atheroscler Rep. 2020. PMID: 32772254 Review.
Cited by
-
CD36 Mediated Fatty Acid-Induced Podocyte Apoptosis via Oxidative Stress.PLoS One. 2015 May 22;10(5):e0127507. doi: 10.1371/journal.pone.0127507. eCollection 2015. PLoS One. 2015. PMID: 26000608 Free PMC article.
-
Lipid Players of Cellular Senescence.Metabolites. 2020 Aug 21;10(9):339. doi: 10.3390/metabo10090339. Metabolites. 2020. PMID: 32839400 Free PMC article. Review.
-
CD36 Signaling in Diabetic Cardiomyopathy.Aging Dis. 2021 Jun 1;12(3):826-840. doi: 10.14336/AD.2020.1217. eCollection 2021 Jun. Aging Dis. 2021. PMID: 34094645 Free PMC article. Review.
-
A CD36-related transmembrane protein is coordinated with an intracellular lipid-binding protein in selective carotenoid transport for cocoon coloration.J Biol Chem. 2010 Mar 5;285(10):7739-51. doi: 10.1074/jbc.M109.074435. Epub 2010 Jan 6. J Biol Chem. 2010. PMID: 20053988 Free PMC article.
-
Identification of CD36 as a new interaction partner of membrane NEU1: potential implication in the pro-atherogenic effects of the elastin receptor complex.Cell Mol Life Sci. 2019 Feb;76(4):791-807. doi: 10.1007/s00018-018-2978-6. Epub 2018 Nov 29. Cell Mol Life Sci. 2019. PMID: 30498996 Free PMC article.
References
-
- Tandon N. N., Kralisz U., Jamieson G. A. ( 1989) J. Biol. Chem. 264, 7576– 7583 - PubMed
-
- Greenwalt D. E., Watt K. W., So O. Y., Jiwani N. ( 1990) Biochemistry 29, 7054– 7059 - PubMed
-
- Endemann G., Stanton L. W., Madden K. S., Bryant C. M., White R. T., Protter A. A. ( 1993) J. Biol. Chem. 268, 11811– 11816 - PubMed
-
- Greenwalt D. E., Lipsky R. H., Ockenhouse C. F., Ikeda H., Tandon N. N., Jamieson G. A. ( 1992) Blood 80, 1105– 1115 - PubMed
-
- Harmon C. M., Abumrad N. A. ( 1993) J. Membr. Biol. 133, 43– 49 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

