Mechanisms of chiral discrimination by topoisomerase IV
- PMID: 19359479
- PMCID: PMC2667371
- DOI: 10.1073/pnas.0900574106
Mechanisms of chiral discrimination by topoisomerase IV
Abstract
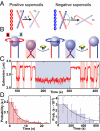
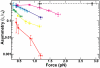
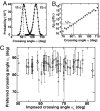
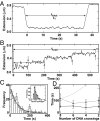
Topoisomerase IV (Topo IV), an essential ATP-dependent bacterial type II topoisomerase, transports one segment of DNA through a transient double-strand break in a second segment of DNA. In vivo, Topo IV unlinks catenated chromosomes before cell division and relaxes positive supercoils generated during DNA replication. In vitro, Topo IV relaxes positive supercoils at least 20-fold faster than negative supercoils. The mechanisms underlying this chiral discrimination by Topo IV and other type II topoisomerases remain speculative. We used magnetic tweezers to measure the relaxation rates of single and multiple DNA crossings by Topo IV. These measurements allowed us to determine unambiguously the relative importance of DNA crossing geometry and enzymatic processivity in chiral discrimination by Topo IV. Our results indicate that Topo IV binds and passes DNA strands juxtaposed in a nearly perpendicular orientation and that relaxation of negative supercoiled DNA is perfectly distributive. Together, these results suggest that chiral discrimination arises primarily from dramatic differences in the processivity of relaxing positive and negative supercoiled DNA: Topo IV is highly processive on positively supercoiled DNA, whereas it is perfectly distributive on negatively supercoiled DNA. These results provide fresh insight into topoisomerase mechanisms and lead to a model that reconciles contradictory aspects of previous findings while providing a framework to interpret future results.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Chiral discrimination and writhe-dependent relaxation mechanism of human topoisomerase IIα.J Biol Chem. 2013 May 10;288(19):13695-703. doi: 10.1074/jbc.M112.444745. Epub 2013 Mar 18. J Biol Chem. 2013. PMID: 23508957 Free PMC article.
-
Chirality sensing by Escherichia coli topoisomerase IV and the mechanism of type II topoisomerases.Proc Natl Acad Sci U S A. 2003 Jul 22;100(15):8654-9. doi: 10.1073/pnas.1133178100. Epub 2003 Jul 11. Proc Natl Acad Sci U S A. 2003. PMID: 12857958 Free PMC article.
-
Alteration of Escherichia coli topoisomerase IV conformation upon enzyme binding to positively supercoiled DNA.J Biol Chem. 2006 Jul 14;281(28):18927-32. doi: 10.1074/jbc.M603068200. Epub 2006 May 9. J Biol Chem. 2006. PMID: 16684778
-
What makes a type IIA topoisomerase a gyrase or a Topo IV?Nucleic Acids Res. 2021 Jun 21;49(11):6027-6042. doi: 10.1093/nar/gkab270. Nucleic Acids Res. 2021. PMID: 33905522 Free PMC article. Review.
-
The mechanism of negative DNA supercoiling: a cascade of DNA-induced conformational changes prepares gyrase for strand passage.DNA Repair (Amst). 2014 Apr;16:23-34. doi: 10.1016/j.dnarep.2014.01.011. Epub 2014 Feb 22. DNA Repair (Amst). 2014. PMID: 24674625 Review.
Cited by
-
Telling Your Right Hand from Your Left: The Effects of DNA Supercoil Handedness on the Actions of Type II Topoisomerases.Int J Mol Sci. 2023 Jul 7;24(13):11199. doi: 10.3390/ijms241311199. Int J Mol Sci. 2023. PMID: 37446377 Free PMC article. Review.
-
The dimer state of GyrB is an active form: implications for the initial complex assembly and processive strand passage.Nucleic Acids Res. 2011 Oct;39(19):8488-502. doi: 10.1093/nar/gkr553. Epub 2011 Jul 10. Nucleic Acids Res. 2011. PMID: 21745817 Free PMC article.
-
Interplay of DNA supercoiling and catenation during the segregation of sister duplexes.Nucleic Acids Res. 2009 Aug;37(15):5126-37. doi: 10.1093/nar/gkp530. Epub 2009 Jun 24. Nucleic Acids Res. 2009. PMID: 19553196 Free PMC article.
-
Supercoiling and looping promote DNA base accessibility and coordination among distant sites.Nat Commun. 2021 Sep 28;12(1):5683. doi: 10.1038/s41467-021-25936-2. Nat Commun. 2021. PMID: 34584096 Free PMC article.
-
The Localization and Action of Topoisomerase IV in Escherichia coli Chromosome Segregation Is Coordinated by the SMC Complex, MukBEF.Cell Rep. 2015 Dec 22;13(11):2587-2596. doi: 10.1016/j.celrep.2015.11.034. Epub 2015 Dec 10. Cell Rep. 2015. PMID: 26686641 Free PMC article.
References
-
- Corbett KD, Berger JM. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu Rev Biophys Biomol Struct. 2004;33:95–118. - PubMed
-
- Wang JC. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–692. - PubMed
-
- Champoux JJ. DNA topoisomerases: Structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. - PubMed
-
- Wang JC. Moving one DNA double helix through another by a type II DNA topoisomerase: The story of a simple molecular machine. Q Rev Biophys. 1998;31:107–144. - PubMed
-
- Espeli O, Marians KJ. Untangling intracellular DNA topology. Mol Microbiol. 2004;52:925–931. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

