Alternative polyadenylation variants of the RNA binding protein, HuR: abundance, role of AU-rich elements and auto-Regulation
- PMID: 19359363
- PMCID: PMC2699509
- DOI: 10.1093/nar/gkp223
Alternative polyadenylation variants of the RNA binding protein, HuR: abundance, role of AU-rich elements and auto-Regulation
Abstract
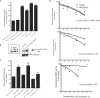
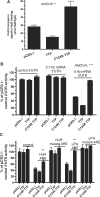
The RNA-binding protein, HuR, is involved in the stabilization of AU-rich element-containing mRNAs with products that are involved in cell-cycle progression, cell differentiation and inflammation. We show that there are multiple polyadenylation variants of HuR mRNA that differ in their abundance, using both bioinformatics and experimental approaches. A polyadenylation variant with distal poly(A) signal is a rare transcript that harbors functional AU-rich elements (ARE) in the 3'UTR. A minimal 60-nt region, but not a mutant form, fused to reporter-3'UTR constructs was able to downregulate the reporter activity. The most predominant and alternatively polyadenylated mature transcript does not contain the ARE. HuR itself binds HuR mRNA, and upregulated the activity of reporter from constructs fused with ARE-isoform and the HuR ARE. Wild-type tristetraprolin (TTP), but not the zinc finger mutant TTP, competes for HuR binding and upregulation of HuR mRNA. The study shows that the HuR gene codes for several polyadenylation variants differentially regulated by AU-rich elements, and demonstrates an auto-regulatory role of HuR.
Figures




Similar articles
-
Interaction between 3' untranslated region of calcitonin receptor messenger ribonucleic acid (RNA) and adenylate/uridylate (AU)-rich element binding proteins (AU-rich RNA-binding factor 1 and Hu antigen R).Endocrinology. 2004 Apr;145(4):1730-8. doi: 10.1210/en.2003-0862. Epub 2004 Jan 8. Endocrinology. 2004. PMID: 14715706
-
Systematic Analysis of AU-Rich Element Expression in Cancer Reveals Common Functional Clusters Regulated by Key RNA-Binding Proteins.Cancer Res. 2016 Jul 15;76(14):4068-80. doi: 10.1158/0008-5472.CAN-15-3110. Epub 2016 May 17. Cancer Res. 2016. PMID: 27197193
-
HADHB, HuR, and CP1 bind to the distal 3'-untranslated region of human renin mRNA and differentially modulate renin expression.J Biol Chem. 2003 Nov 7;278(45):44894-903. doi: 10.1074/jbc.M307782200. Epub 2003 Aug 21. J Biol Chem. 2003. PMID: 12933794
-
HuR and mRNA stability.Cell Mol Life Sci. 2001 Feb;58(2):266-77. doi: 10.1007/PL00000854. Cell Mol Life Sci. 2001. PMID: 11289308 Free PMC article. Review.
-
Multiple functions of tristetraprolin/TIS11 RNA-binding proteins in the regulation of mRNA biogenesis and degradation.Cell Mol Life Sci. 2013 Jun;70(12):2031-44. doi: 10.1007/s00018-012-1150-y. Epub 2012 Sep 12. Cell Mol Life Sci. 2013. PMID: 22968342 Free PMC article. Review.
Cited by
-
miR-29a inhibition normalizes HuR over-expression and aberrant AU-rich mRNA stability in invasive cancer.J Pathol. 2013 May;230(1):28-38. doi: 10.1002/path.4178. Epub 2013 Mar 21. J Pathol. 2013. PMID: 23401122 Free PMC article.
-
p16( INK4a) positively regulates cyclin D1 and E2F1 through negative control of AUF1.PLoS One. 2011;6(7):e21111. doi: 10.1371/journal.pone.0021111. Epub 2011 Jul 20. PLoS One. 2011. PMID: 21799732 Free PMC article.
-
Hyper conserved elements in vertebrate mRNA 3'-UTRs reveal a translational network of RNA-binding proteins controlled by HuR.Nucleic Acids Res. 2013 Mar 1;41(5):3201-16. doi: 10.1093/nar/gkt017. Epub 2013 Feb 1. Nucleic Acids Res. 2013. PMID: 23376935 Free PMC article.
-
Post-transcriptional regulation of BIRC5/survivin expression and induction of apoptosis in breast cancer cells by tristetraprolin.RNA Biol. 2024 Jan;21(1):1-15. doi: 10.1080/15476286.2023.2286101. Epub 2023 Dec 18. RNA Biol. 2024. PMID: 38111129 Free PMC article.
-
Alternative polyadenylation dependent function of splicing factor SRSF3 contributes to cellular senescence.Aging (Albany NY). 2019 Mar 4;11(5):1356-1388. doi: 10.18632/aging.101836. Aging (Albany NY). 2019. PMID: 30835716 Free PMC article.
References
-
- Blackshear PJ. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem. Soc. Trans. 2002;30:945–952. - PubMed
-
- Ma WJ, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 1996;271:8144–8151. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

