Amphipathic small molecules mimic the binding mode and function of endogenous transcription factors
- PMID: 19348463
- PMCID: PMC2744096
- DOI: 10.1021/cb900028j
Amphipathic small molecules mimic the binding mode and function of endogenous transcription factors
Abstract
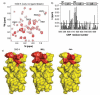
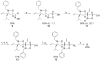
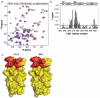
Small molecules that reconstitute the binding mode(s) of a protein and in doing so elicit a programmed functional response offer considerable advantages in the control of complex biological processes. The development challenges of such molecules are significant, however. Many protein-protein interactions require multiple points of contact over relatively large surface areas. More significantly, several binding modes can be superimposed upon a single sequence within a protein, and a true small molecule replacement must be preprogrammed for such multimodal binding. This is the case for the transcriptional activation domain or TAD of transcriptional activators as these motifs utilize a poorly characterized multipartner binding profile in order to stimulate gene expression. Here we describe a unique class of small molecules that exhibit both function and a binding profile analogous to natural transcriptional activation domains. Of particular note, the small molecules are the first reported to bind to the KIX domain within the CREB binding protein (CBP) at a site that is utilized by natural activators. Further, a comparison of functional and nonfunctional small molecules indicates that an interaction with CBP is a key contributor to transcriptional activity. Taken together, the evidence suggests that the small molecule TADs mimic both the function and mechanism of their natural counterparts and thus present a framework for the broader development of small molecule transcriptional switches.
Figures
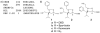
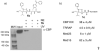
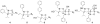

Similar articles
-
Cooperativity in transcription factor binding to the coactivator CREB-binding protein (CBP). The mixed lineage leukemia protein (MLL) activation domain binds to an allosteric site on the KIX domain.J Biol Chem. 2002 Nov 8;277(45):43168-74. doi: 10.1074/jbc.M207660200. Epub 2002 Aug 29. J Biol Chem. 2002. PMID: 12205094
-
Transcriptional tools: Small molecules for modulating CBP KIX-dependent transcriptional activators.Biopolymers. 2011 Jan;95(1):17-23. doi: 10.1002/bip.21548. Biopolymers. 2011. PMID: 20882601 Free PMC article.
-
Structural and mechanistic insights into the interaction of the circadian transcription factor BMAL1 with the KIX domain of the CREB-binding protein.J Biol Chem. 2019 Nov 8;294(45):16604-16619. doi: 10.1074/jbc.RA119.009845. Epub 2019 Sep 12. J Biol Chem. 2019. PMID: 31515273 Free PMC article.
-
Structurally distinct modes of recognition of the KIX domain of CBP by Jun and CREB.Biochemistry. 2002 Nov 26;41(47):13956-64. doi: 10.1021/bi026222m. Biochemistry. 2002. PMID: 12437352
-
Role of co-activators and co-repressors in the mechanism of steroid/thyroid receptor action.Recent Prog Horm Res. 1997;52:141-64; discussion 164-5. Recent Prog Horm Res. 1997. PMID: 9238851 Review.
Cited by
-
Inhibition of CREB Binding and Function with a Dual-Targeting Ligand.Biochemistry. 2024 Jan 2;63(1):1-8. doi: 10.1021/acs.biochem.3c00469. Epub 2023 Dec 12. Biochemistry. 2024. PMID: 38086054 Free PMC article.
-
Profiling the dynamic interfaces of fluorinated transcription complexes for ligand discovery and characterization.ACS Chem Biol. 2012 Aug 17;7(8):1345-50. doi: 10.1021/cb3002733. Epub 2012 Jul 2. ACS Chem Biol. 2012. PMID: 22725662 Free PMC article.
-
Assessing helical protein interfaces for inhibitor design.J Am Chem Soc. 2011 Sep 14;133(36):14220-3. doi: 10.1021/ja206074j. Epub 2011 Aug 22. J Am Chem Soc. 2011. PMID: 21846146 Free PMC article.
-
Unexpected specificity within dynamic transcriptional protein-protein complexes.Proc Natl Acad Sci U S A. 2020 Nov 3;117(44):27346-27353. doi: 10.1073/pnas.2013244117. Epub 2020 Oct 19. Proc Natl Acad Sci U S A. 2020. PMID: 33077600 Free PMC article.
-
Structure, function and inhibition of critical protein-protein interactions involving mixed lineage leukemia 1 and its fusion oncoproteins.J Hematol Oncol. 2021 Apr 6;14(1):56. doi: 10.1186/s13045-021-01057-7. J Hematol Oncol. 2021. PMID: 33823889 Free PMC article. Review.
References
-
- Ptashne M, Gann A. Genes & Signals. Cold Spring Harbor Laboratory; New York: 2001.
-
- Mapp AK, Ansari AZ. A TAD further: exogenous control of gene activation. ACS Chem Biol. 2007;2:62–75. - PubMed
-
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Aksien LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. - PubMed
-
- Darnell JE. Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2:740–749. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

