Bile acids: regulation of synthesis
- PMID: 19346330
- PMCID: PMC2739756
- DOI: 10.1194/jlr.R900010-JLR200
Bile acids: regulation of synthesis
Abstract
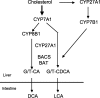
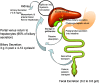
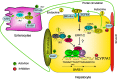
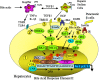
Bile acids are physiological detergents that generate bile flow and facilitate intestinal absorption and transport of lipids, nutrients, and vitamins. Bile acids also are signaling molecules and inflammatory agents that rapidly activate nuclear receptors and cell signaling pathways that regulate lipid, glucose, and energy metabolism. The enterohepatic circulation of bile acids exerts important physiological functions not only in feedback inhibition of bile acid synthesis but also in control of whole-body lipid homeostasis. In the liver, bile acids activate a nuclear receptor, farnesoid X receptor (FXR), that induces an atypical nuclear receptor small heterodimer partner, which subsequently inhibits nuclear receptors, liver-related homolog-1, and hepatocyte nuclear factor 4alpha and results in inhibiting transcription of the critical regulatory gene in bile acid synthesis, cholesterol 7alpha-hydroxylase (CYP7A1). In the intestine, FXR induces an intestinal hormone, fibroblast growth factor 15 (FGF15; or FGF19 in human), which activates hepatic FGF receptor 4 (FGFR4) signaling to inhibit bile acid synthesis. However, the mechanism by which FXR/FGF19/FGFR4 signaling inhibits CYP7A1 remains unknown. Bile acids are able to induce FGF19 in human hepatocytes, and the FGF19 autocrine pathway may exist in the human livers. Bile acids and bile acid receptors are therapeutic targets for development of drugs for treatment of cholestatic liver diseases, fatty liver diseases, diabetes, obesity, and metabolic syndrome.
Figures
Similar articles
-
Bile acids activate fibroblast growth factor 19 signaling in human hepatocytes to inhibit cholesterol 7alpha-hydroxylase gene expression.Hepatology. 2009 Jan;49(1):297-305. doi: 10.1002/hep.22627. Hepatology. 2009. PMID: 19085950 Free PMC article.
-
Fibroblast Growth Factor 21 Response in a Preclinical Alcohol Model of Acute-on-Chronic Liver Injury.Int J Mol Sci. 2021 Jul 23;22(15):7898. doi: 10.3390/ijms22157898. Int J Mol Sci. 2021. PMID: 34360670 Free PMC article.
-
FXR an emerging therapeutic target for the treatment of atherosclerosis.J Cell Mol Med. 2010 Jan;14(1-2):79-92. doi: 10.1111/j.1582-4934.2009.00997.x. J Cell Mol Med. 2010. PMID: 20041971 Free PMC article. Review.
-
Bile Acids as Hormones: The FXR-FGF15/19 Pathway.Dig Dis. 2015;33(3):327-31. doi: 10.1159/000371670. Epub 2015 May 27. Dig Dis. 2015. PMID: 26045265 Free PMC article. Review.
-
Molecular Basis of Bile Acid-FXR-FGF15/19 Signaling Axis.Int J Mol Sci. 2022 May 27;23(11):6046. doi: 10.3390/ijms23116046. Int J Mol Sci. 2022. PMID: 35682726 Free PMC article. Review.
Cited by
-
Regulation of cholesterol and bile acid homeostasis by the cholesterol 7α-hydroxylase/steroid response element-binding protein 2/microRNA-33a axis in mice.Hepatology. 2013 Sep;58(3):1111-21. doi: 10.1002/hep.26427. Epub 2013 Jul 31. Hepatology. 2013. PMID: 23536474 Free PMC article.
-
Intestinal synthesis and secretion of bile salts as an adaptation to developmental biliary atresia in the sea lamprey.Proc Natl Acad Sci U S A. 2012 Jul 10;109(28):11419-24. doi: 10.1073/pnas.1203008109. Epub 2012 Jun 25. Proc Natl Acad Sci U S A. 2012. PMID: 22733776 Free PMC article.
-
Transcriptome analysis of the liver of Eospalax fontanierii under hypoxia.PeerJ. 2021 Apr 22;9:e11166. doi: 10.7717/peerj.11166. eCollection 2021. PeerJ. 2021. PMID: 33981491 Free PMC article.
-
Glucose and insulin induction of bile acid synthesis: mechanisms and implication in diabetes and obesity.J Biol Chem. 2012 Jan 13;287(3):1861-73. doi: 10.1074/jbc.M111.305789. Epub 2011 Dec 5. J Biol Chem. 2012. PMID: 22144677 Free PMC article.
-
Xenobiotic and Endobiotic Mediated Interactions Between the Cytochrome P450 System and the Inflammatory Response in the Liver.Adv Pharmacol. 2015;74:131-61. doi: 10.1016/bs.apha.2015.04.001. Epub 2015 May 19. Adv Pharmacol. 2015. PMID: 26233906 Free PMC article. Review.
References
-
- Myant N. B., Mitropoulos K. A. 1977. Cholesterol 7α-hydroxylase. J. Lipid Res. 18: 135–153. - PubMed
-
- Noshiro M., Nishimoto M., Morohashi K., Okuda K. 1989. Molecular cloning of cDNA for cholesterol 7α-hydroxylase from rat liver microsomes. Nucleotide sequence and expression. FEBS Lett. 257: 97–100. - PubMed
-
- Li Y. C., Wang D. P., Chiang J. Y. 1990. Regulation of cholesterol 7α-hydroxylase in the liver. Cloning, sequencing, and regulation of cholesterol 7α-hydroxylase mRNA. J. Biol. Chem. 265: 12012–12019. - PubMed
-
- Makishima M., Okamoto A. Y., Repa J. J., Tu H., Learned R. M., Luk A., Hull M. V., Lustig K. D., Mangelsdorf D. J., Shan B. 1999. Identification of a nuclear receptor for bile acids. Science. 284: 1362–1365. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

