Acetylation targets mutant huntingtin to autophagosomes for degradation
- PMID: 19345187
- PMCID: PMC2940108
- DOI: 10.1016/j.cell.2009.03.018
Acetylation targets mutant huntingtin to autophagosomes for degradation
Abstract
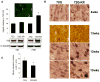
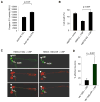
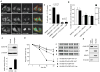
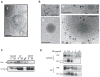
Huntington's disease (HD) is an incurable neurodegenerative disease caused by neuronal accumulation of the mutant protein huntingtin. Improving clearance of the mutant protein is expected to prevent cellular dysfunction and neurodegeneration in HD. We report here that such clearance can be achieved by posttranslational modification of the mutant Huntingtin (Htt) by acetylation at lysine residue 444 (K444). Increased acetylation at K444 facilitates trafficking of mutant Htt into autophagosomes, significantly improves clearance of the mutant protein by macroautophagy, and reverses the toxic effects of mutant huntingtin in primary striatal and cortical neurons and in a transgenic C. elegans model of HD. In contrast, mutant Htt that is rendered resistant to acetylation dramatically accumulates and leads to neurodegeneration in cultured neurons and in mouse brain. These studies identify acetylation as a mechanism for removing accumulated protein in HD, and more broadly for actively targeting proteins for degradation by autophagy.
Figures



Similar articles
-
The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation.J Neurosci. 2014 Jan 22;34(4):1293-305. doi: 10.1523/JNEUROSCI.1870-13.2014. J Neurosci. 2014. PMID: 24453320 Free PMC article.
-
PRMT5- mediated symmetric arginine dimethylation is attenuated by mutant huntingtin and is impaired in Huntington's disease (HD).Cell Cycle. 2015;14(11):1716-29. doi: 10.1080/15384101.2015.1033595. Cell Cycle. 2015. PMID: 25927346 Free PMC article.
-
Mass spectrometric identification of novel lysine acetylation sites in huntingtin.Mol Cell Proteomics. 2011 Oct;10(10):M111.009829. doi: 10.1074/mcp.M111.009829. Epub 2011 Jun 18. Mol Cell Proteomics. 2011. PMID: 21685499 Free PMC article.
-
Small changes, big impact: posttranslational modifications and function of huntingtin in Huntington disease.Neuroscientist. 2011 Oct;17(5):475-92. doi: 10.1177/1073858410390378. Epub 2011 Feb 10. Neuroscientist. 2011. PMID: 21311053 Free PMC article. Review.
-
Huntingtin processing in pathogenesis of Huntington disease.Acta Pharmacol Sin. 2004 Oct;25(10):1243-9. Acta Pharmacol Sin. 2004. PMID: 15456523 Review.
Cited by
-
Macroautophagy abnormality in essential tremor.PLoS One. 2012;7(12):e53040. doi: 10.1371/journal.pone.0053040. Epub 2012 Dec 27. PLoS One. 2012. PMID: 23300858 Free PMC article.
-
Identification of natural products with neuronal and metabolic benefits through autophagy induction.Autophagy. 2017 Jan 2;13(1):41-56. doi: 10.1080/15548627.2016.1240855. Epub 2016 Oct 28. Autophagy. 2017. PMID: 27791467 Free PMC article.
-
Targeting histone deacetylases for the treatment of Huntington's disease.CNS Neurosci Ther. 2010 Dec;16(6):348-61. doi: 10.1111/j.1755-5949.2010.00184.x. CNS Neurosci Ther. 2010. PMID: 20642797 Free PMC article. Review.
-
Protein modifications involved in neurotransmitter and gasotransmitter signaling.Trends Neurosci. 2010 Nov;33(11):493-502. doi: 10.1016/j.tins.2010.07.004. Epub 2010 Sep 16. Trends Neurosci. 2010. PMID: 20843563 Free PMC article. Review.
-
Dysregulation of histone deacetylases in carcinogenesis and tumor progression: a possible link to apoptosis and autophagy.Cell Mol Life Sci. 2019 Sep;76(17):3263-3282. doi: 10.1007/s00018-019-03098-1. Epub 2019 Apr 13. Cell Mol Life Sci. 2019. PMID: 30982077 Free PMC article. Review.
References
-
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. - PubMed
-
- Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. - PubMed
-
- Cuervo AM. Autophagy: many paths to the same end. Mol Cell Biochem. 2004;263:55–72. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

